

| Basic Information | |
|---|---|
| Species | Arabidopsis thaliana |
| Cazyme ID | AT2G15350.1 |
| Family | GT37 |
| Protein Properties | Length: 441 Molecular Weight: 50837.1 Isoelectric Point: 6.6218 |
| Chromosome | Chromosome/Scaffold: 2 Start: 6688140 End: 6689462 |
| Description | xyloglucan fucosyltransferase, putative, expressed |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT37 | 2 | 410 | 0 |
| PSEYLVSELRSYEMLHKRCGPDTKAYKEATEKLSRDEYYASESNGECRYIVWLARDGLGNRLITLASVFLYAILTERIILVDNRKDVSDLLCEPFPGTSW LLPLDFPMLNYTYAYGYNKEYPRCYGTMLENHAINSTSIPPHLYLHNIHDSRDSDKLFFCQKDQSFIDKVPWLIIQTNAYFVPSLWLNPTFQTKLVKLFP QKETVFHHLARYLFHPTNEVWDMVTKYYDAHLSNADERLGIQIRVFGKPSGYFKHVMDQVVACTQREKLLPEFEEESKVNISKPPKLKVVLVASLYPEYS VNLTNMFLARPSSTGEIIEVYQPSAERVQQTDKKSHDQKALAEMYLLSLTDNIVTSGWSTFGYVSYSLGGLKPWLLYQPVNFTTPNPPCVRSKSMEPCYH TPPSHGCEA | |||
| Full Sequence |
|---|
| Protein Sequence Length: 441 Download |
| MPSEYLVSEL RSYEMLHKRC GPDTKAYKEA TEKLSRDEYY ASESNGECRY IVWLARDGLG 60 NRLITLASVF LYAILTERII LVDNRKDVSD LLCEPFPGTS WLLPLDFPML NYTYAYGYNK 120 EYPRCYGTML ENHAINSTSI PPHLYLHNIH DSRDSDKLFF CQKDQSFIDK VPWLIIQTNA 180 YFVPSLWLNP TFQTKLVKLF PQKETVFHHL ARYLFHPTNE VWDMVTKYYD AHLSNADERL 240 GIQIRVFGKP SGYFKHVMDQ VVACTQREKL LPEFEEESKV NISKPPKLKV VLVASLYPEY 300 SVNLTNMFLA RPSSTGEIIE VYQPSAERVQ QTDKKSHDQK ALAEMYLLSL TDNIVTSGWS 360 TFGYVSYSLG GLKPWLLYQP VNFTTPNPPC VRSKSMEPCY HTPPSHGCEA DWGTNSGKIL 420 PFVRHCEDMM YGGLKLYDDF * |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd11548 | NodZ_like | 1.0e-6 | 51 | 371 | 333 | + Alpha 1,6-fucosyltransferase similar to Bradyrhizobium NodZ. Bradyrhizobium NodZ is an alpha 1,6-fucosyltransferase involved in the biosynthesis of the nodulation factor, a lipo-chitooligosaccharide formed by three-to-six beta-1,4-linked N-acetyl-d-glucosamine (GlcNAc) residues and a fatty acid acyl group attached to the nitrogen atom at the non-reducing end. NodZ transfers L-fucose from the GDP-beta-L-fucose donor to the reducing residue of the chitin oligosaccharide backbone, before the attachment of a fatty acid group. O-fucosyltransferase-like proteins are GDP-fucose dependent enzymes with similarities to the family 1 glycosyltransferases (GT1). They are soluble ER proteins that may be proteolytically cleaved from a membrane-associated preprotein, and are involved in the O-fucosylation of protein substrates, the core fucosylation of growth factor receptors, and other processes. | ||
| pfam03254 | XG_FTase | 0 | 2 | 410 | 414 | + Xyloglucan fucosyltransferase. Plant cell walls are crucial for development, signal transduction, and disease resistance in plants. Cell walls are made of cellulose, hemicelluloses, and pectins. Xyloglucan (XG), the principal load-bearing hemicellulose of dicotyledonous plants, has a terminal fucosyl residue. This fucosyltransferase adds this residue. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity |
| GO:0008417 | fucosyltransferase activity |
| GO:0016020 | membrane |
| GO:0016757 | transferase activity, transferring glycosyl groups |
| GO:0042546 | cell wall biogenesis |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAL50624.1 | 0 | 2 | 440 | 77 | 515 | AF417475_1 fucosyltransferase-like protein FUT5 [Arabidopsis thaliana] |
| RefSeq | NP_179137.1 | 0 | 1 | 440 | 1 | 440 | FUT10 (FUCOSYLTRANSFERASE 10); fucosyltransferase/ transferase, transferring glycosyl groups [Arabidopsis thaliana] |
| RefSeq | NP_179139.1 | 0 | 2 | 440 | 95 | 533 | FUT5; fucosyltransferase/ transferase, transferring glycosyl groups [Arabidopsis thaliana] |
| RefSeq | NP_179141.1 | 0 | 2 | 440 | 65 | 503 | FUT4; fucosyltransferase/ transferase, transferring glycosyl groups [Arabidopsis thaliana] |
| Swiss-Prot | Q9SJP6 | 0 | 1 | 440 | 75 | 514 | FUT10_ARATH RecName: Full=Putative fucosyltransferase 10; Short=AtFUT10 |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2wg4_B | 0.0003 | 51 | 151 | 132 | 218 | A Chain A, Crystal Structure Of The Complex Between Human Hedgehog- Interacting Protein Hip And Sonic Hedgehog Without Calcium |
| PDB | 2wfx_B | 0.0003 | 51 | 151 | 132 | 218 | B Chain B, Crystal Structure Of The Complex Between Human Hedgehog- Interacting Protein Hip And Sonic Hedgehog In The Presence Of Calcium |
| PDB | 2wg3_D | 0.0003 | 51 | 151 | 132 | 218 | C Chain C, Crystal Structure Of The Complex Between Human Hedgehog- Interacting Protein Hip And Desert Hedgehog Without Calcium |
| PDB | 2wg3_C | 0.0003 | 51 | 151 | 132 | 218 | C Chain C, Crystal Structure Of The Complex Between Human Hedgehog- Interacting Protein Hip And Desert Hedgehog Without Calcium |
| PDB | 2wft_B | 0.0003 | 51 | 151 | 132 | 218 | A Chain A, Crystal Structure Of The Human Hip Ectodomain |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
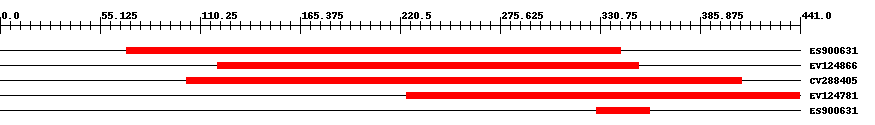 | ||||
| Hit | Length | Start | End | EValue |
| ES900631 | 273 | 70 | 342 | 0 |
| EV124866 | 235 | 120 | 352 | 0 |
| CV288405 | 308 | 103 | 409 | 0 |
| EV124781 | 220 | 224 | 441 | 0 |
| ES900631 | 30 | 329 | 358 | 2 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |