

| Basic Information | |
|---|---|
| Species | Arabidopsis thaliana |
| Cazyme ID | AT3G22250.1 |
| Family | GT1 |
| Protein Properties | Length: 462 Molecular Weight: 52145.8 Isoelectric Point: 6.5177 |
| Chromosome | Chromosome/Scaffold: 3 Start: 7867753 End: 7870229 |
| Description | UDP-glucoronosyl/UDP-glucosyl transferase family protein, putative, expressed |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT1 | 274 | 441 | 9.6e-33 |
| LGWLQEQNPNSVIYISFGSWVSPIGESNIQTLALALEASGRPFLWALNRVWQEGLPPGFVHRVTITKNQGRIVSWAPQLEVLRNDSVGCYVTHCGWNSTM EAVASSRRLLCYPVAGDQFVNCKYIVDVWKIGVRLSGFGEKEVEDGLRKVMEDQDMGERLRKLRDRAM | |||
| Full Sequence |
|---|
| Protein Sequence Length: 462 Download |
| MKVTQKPKII FIPYPAQGHV TPMLHLASAF LSRGFSPVVM TPESIHRRIS ATNEDLGITF 60 LALSDGQDRP DAPPSDFFSI ENSMENIMPP QLERLLLEED LDVACVVVDL LASWAIGVAD 120 RCGVPVAGFW PVMFAAYRLI QAIPELVRTG LVSQKGCPRQ LEKTIVQPEQ PLLSAEDLPW 180 LIGTPKAQKK RFKFWQRTLE RTKSLRWILT SSFKDEYEDV DNHKASYKKS NDLNKENNGQ 240 NPQILHLGPL HNQEATNNIT ITKTSFWEED MSCLGWLQEQ NPNSVIYISF GSWVSPIGES 300 NIQTLALALE ASGRPFLWAL NRVWQEGLPP GFVHRVTITK NQGRIVSWAP QLEVLRNDSV 360 GCYVTHCGWN STMEAVASSR RLLCYPVAGD QFVNCKYIVD VWKIGVRLSG FGEKEVEDGL 420 RKVMEDQDMG ERLRKLRDRA MGNEARLSSE MNFTFLKNEL N* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02448 | PLN02448 | 3.0e-35 | 12 | 438 | 475 | + UDP-glycosyltransferase family protein | ||
| PLN02173 | PLN02173 | 2.0e-38 | 9 | 441 | 456 | + UDP-glucosyl transferase family protein | ||
| PLN02410 | PLN02410 | 1.0e-39 | 8 | 435 | 439 | + UDP-glucoronosyl/UDP-glucosyl transferase family protein | ||
| PLN02555 | PLN02555 | 1.0e-45 | 1 | 460 | 501 | + limonoid glucosyltransferase | ||
| PLN02562 | PLN02562 | 0 | 1 | 461 | 465 | + UDP-glycosyltransferase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0008152 | metabolic process |
| GO:0008194 | UDP-glycosyltransferase activity |
| GO:0016757 | transferase activity, transferring glycosyl groups |
| GO:0016758 | transferase activity, transferring hexosyl groups |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| DDBJ | BAD44605.1 | 0 | 1 | 461 | 1 | 461 | hypothetical protein [Arabidopsis thaliana] |
| EMBL | CBI27396.1 | 0 | 1 | 450 | 1 | 435 | unnamed protein product [Vitis vinifera] |
| RefSeq | NP_188864.1 | 0 | 1 | 461 | 1 | 461 | UDP-glucoronosyl/UDP-glucosyl transferase family protein [Arabidopsis thaliana] |
| RefSeq | XP_002266304.1 | 0 | 1 | 461 | 1 | 446 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002521042.1 | 0 | 1 | 460 | 1 | 448 | UDP-glucosyltransferase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2pq6_A | 8e-40 | 5 | 441 | 7 | 450 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt85h2- Insights Into The Structural Basis Of A Multifunctional (Iso) Flavonoid Glycosyltransferase |
| PDB | 2c9z_A | 3e-37 | 103 | 456 | 113 | 446 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt85h2- Insights Into The Structural Basis Of A Multifunctional (Iso) Flavonoid Glycosyltransferase |
| PDB | 2c1z_A | 3e-37 | 103 | 456 | 113 | 446 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt85h2- Insights Into The Structural Basis Of A Multifunctional (Iso) Flavonoid Glycosyltransferase |
| PDB | 2c1x_A | 3e-37 | 103 | 456 | 113 | 446 | A Chain A, Structure And Activity Of A Flavonoid 3-O Glucosyltransferase Reveals The Basis For Plant Natural Product Modification |
| PDB | 3hbj_A | 2e-31 | 9 | 456 | 16 | 448 | A Chain A, Structure And Activity Of A Flavonoid 3-O Glucosyltransferase Reveals The Basis For Plant Natural Product Modification |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| cytokinins-O-glucoside biosynthesis | RXN-4723 | EC-2.4.1.203 | trans-zeatin O-β-D-glucosyltransferase |
| cytokinins-O-glucoside biosynthesis | RXN-4726 | EC-2.4.1 | UDP-glucose:dihydrozeatin O-β-D-glucosyltransferase |
| cytokinins-O-glucoside biosynthesis | RXN-4735 | EC-2.4.1.215 | cis-zeatin O-β-D-glucosyltransferase |
| cytokinins-O-glucoside biosynthesis | RXN-4737 | EC-2.4.1 | dihydrozeatin-9-N-glucoside O-β-D-glucosyltransferase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
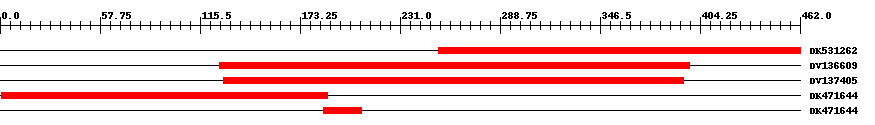 | ||||
| Hit | Length | Start | End | EValue |
| DK531262 | 211 | 253 | 462 | 0 |
| DV136609 | 272 | 127 | 398 | 0 |
| DV137405 | 267 | 129 | 395 | 0 |
| DK471644 | 189 | 1 | 189 | 0 |
| DK471644 | 23 | 187 | 209 | 0.00002 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |