

| Basic Information | |
|---|---|
| Species | Zea mays |
| Cazyme ID | GRMZM2G135966_T01 |
| Family | GH47 |
| Protein Properties | Length: 709 Molecular Weight: 79624.3 Isoelectric Point: 4.4648 |
| Chromosome | Chromosome/Scaffold: 7 Start: 12320495 End: 12329855 |
| Description | alpha-mannosidase 1 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH47 | 577 | 684 | 0 |
| AKEDMSVGTSWNILRPETVESLMYLWRLTGNTTYQDWGWDIFQAFEKNFRVEFGYVGLRDVNTGEKDNMMQSFFLAETLKYLYLLFFPPSVISFEDWVFN TEAHPLRI | |||
| Full Sequence |
|---|
| Protein Sequence Length: 709 Download |
| MELLLQGWIT SAQCMSITPV REELGVDVDT LEGGRREVAV RSPIEDREEA AAFADPSIAR 60 EEGSWGAGGG VAGLAAPKCG FPGVPINFEI DLYIIPRLFE LNSKVLLHFF YSRLVPLVLL 120 LVEGSTPIDI GEHGWEMLLV VKKATQEAGS KFELLGFAAV HNFYHYPESI RLRISQILVL 180 PPYQGEGHGL GLLEAINYIA QSENIYDVTI ESPSDYLQYD HKSMDNFRAC IYDLMKGEIL 240 GSASGTNRKR LLQMPTSFNK EASFVVYWTH EIGDEDEQTV EQQPEYLRTQ EQQLNELVDI 300 QIEEIDGVAK NVTSCGKDKM AELAVGWVVE PNGTTYLKEG TCLIKQPGDT YDDFASSKPD 360 NECHYVVFDL DLNNFYDCGS SGVSLSAFAF LFSELVQYNQ TKVDNITELE RSHARTIDAH 420 TNRNTIASVV AHFGASWCVM SLSMNYKFEE MAQTHLEVLF LYMDADDVQE MVLFILFAVS 480 ISFIHSTLYQ SKASMNICSR SGSRGIKLTI QMWETSMEGQ LSLTKKTTPS NYYYICEKNG 540 GSLSDKMDEL ACFAPDMLAL GASGYGPEKS EQIMNMAKED MSVGTSWNIL RPETVESLMY 600 LWRLTGNTTY QDWGWDIFQA FEKNFRVEFG YVGLRDVNTG EKDNMMQSFF LAETLKYLYL 660 LFFPPSVISF EDWVFNTEAH PLRIAPVNGN KGIGTPVRPF GRKQGKPE* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
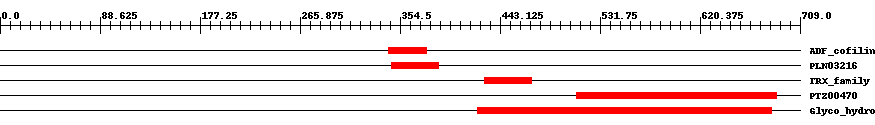 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd11286 | ADF_cofilin_like | 0.0004 | 344 | 378 | 35 | + Cofilin, Destrin, and related actin depolymerizing factors. Actin depolymerization factor/cofilin-like domains (ADF domains) are present in a family of essential eukaryotic actin regulatory proteins. These proteins enhance the turnover rate of actin, and interact with actin monomers (G-actin) as well as actin filaments (F-actin), typically with a preference for ADP-G-actin subunits. The basic function of cofilin is to promote disassembly of aged actin filaments. Vertebrates have three isoforms of cofilin: cofilin-1 (Cfl1, non-muscle cofilin), cofilin-2 (muscle cofilin), and ADF (destrin). When bound to actin monomers, cofilins inhibit their spontaneous exchange of nucleotides. The cooperative binding to (aged) ADP-F-actin induces a local change in the actin filament structure and further promotes aging. | ||
| PLN03216 | PLN03216 | 0.0002 | 347 | 389 | 43 | + actin depolymerizing factor; Provisional | ||
| cd02947 | TRX_family | 4.0e-5 | 429 | 471 | 43 | + TRX family; composed of two groups: Group I, which includes proteins that exclusively encode a TRX domain; and Group II, which are composed of fusion proteins of TRX and additional domains. Group I TRX is a small ancient protein that alter the redox state of target proteins via the reversible oxidation of an active site dithiol, present in a CXXC motif, partially exposed at the protein's surface. TRX reduces protein disulfide bonds, resulting in a disulfide bond at its active site. Oxidized TRX is converted to the active form by TRX reductase, using reducing equivalents derived from either NADPH or ferredoxins. By altering their redox state, TRX regulates the functions of at least 30 target proteins, some of which are enzymes and transcription factors. It also plays an important role in the defense against oxidative stress by directly reducing hydrogen peroxide and certain radicals, and by serving as a reductant for peroxiredoxins. At least two major types of functional TRXs have been reported in most organisms; in eukaryotes, they are located in the cytoplasm and the mitochondria. Higher plants contain more types (at least 20 TRX genes have been detected in the genome of Arabidopsis thaliana), two of which (types f amd m) are located in the same compartment, the chloroplast. Also included in the alignment are TRX-like domains which show sequence homology to TRX but do not contain the redox active CXXC motif. Group II proteins, in addition to either a redox active TRX or a TRX-like domain, also contain additional domains, which may or may not possess homology to known proteins. | ||
| PTZ00470 | PTZ00470 | 1.0e-60 | 511 | 688 | 213 | + glycoside hydrolase family 47 protein; Provisional | ||
| pfam01532 | Glyco_hydro_47 | 7.0e-66 | 423 | 684 | 330 | + Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity |
| GO:0005509 | calcium ion binding |
| GO:0016020 | membrane |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ACN34773.1 | 0 | 506 | 708 | 336 | 565 | unknown [Zea mays] |
| EMBL | CAH66776.1 | 0 | 506 | 708 | 336 | 572 | OSIGBa0113I13.2 [Oryza sativa (indica cultivar-group)] |
| RefSeq | NP_001053800.1 | 0 | 506 | 708 | 336 | 572 | Os04g0606400 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | XP_002448484.1 | 0 | 506 | 708 | 335 | 564 | hypothetical protein SORBIDRAFT_06g027810 [Sorghum bicolor] |
| RefSeq | XP_002528166.1 | 0 | 506 | 706 | 289 | 521 | mannosyl-oligosaccharide alpha-1,2-mannosidase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1nxc_A | 6e-32 | 534 | 684 | 283 | 465 | A Chain A, Structure Of Mouse Golgi Alpha-1,2-Mannosidase Ia Reveals The Molecular Basis For Substrate Specificity Among Class I Enzymes (Family 47 Glycosidases) |
| PDB | 1fo3_A | 4e-30 | 485 | 684 | 207 | 456 | A Chain A, Structure Of Mouse Golgi Alpha-1,2-Mannosidase Ia Reveals The Molecular Basis For Substrate Specificity Among Class I Enzymes (Family 47 Glycosidases) |
| PDB | 1fo2_A | 4e-30 | 485 | 684 | 207 | 456 | A Chain A, Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With 1-Deoxymannojirimycin |
| PDB | 1fmi_A | 4e-30 | 485 | 684 | 207 | 456 | A Chain A, Crystal Structure Of Human Class I Alpha1,2-Mannosidase |
| PDB | 1x9d_A | 6e-30 | 485 | 684 | 285 | 534 | A Chain A, Crystal Structure Of Human Class I Alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue |