

| Basic Information | |
|---|---|
| Species | Zea mays |
| Cazyme ID | GRMZM2G337242_T01 |
| Family | GH38 |
| Protein Properties | Length: 682 Molecular Weight: 75350.1 Isoelectric Point: 7.3167 |
| Chromosome | Chromosome/Scaffold: 1 Start: 117155140 End: 117165079 |
| Description | Glycosyl hydrolase family 38 protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH38 | 51 | 363 | 0 |
| NVHLVPHSHDDVGWLMTIDQYYVGSNNSIQGACVMNTLDSVVDALAKDPARKFIVVEQAFFQRWWAQKSLTVQATVHKLVDSGQLEFINGGWCMHDEAVV HYIDMIDQTTLGHRVIKKQFKKIPRAGWQIDPFGHSAVQAYLLGAELGFDSVHFARIDYQDRATRKANKGLEVIWRGSRPVGSSSQIFTNVFPVHYSAPV GFSFEVLAENVIPVQDDMSLFDYNVQERVDDFVAAAIAQANVTRTNHIMWTMGDDFNYQYAESWFRNMDKLIQYVNKDGRVHALYSTPSIYTDAKHASNE SWPVKYDDYFPYA | |||
| Full Sequence |
|---|
| Protein Sequence Length: 682 Download |
| MAPTLLLVAL VALAVAAATP MVMEASAFTE GCNNRSTPGG GGGTAVGNKL NVHLVPHSHD 60 DVGWLMTIDQ YYVGSNNSIQ GACVMNTLDS VVDALAKDPA RKFIVVEQAF FQRWWAQKSL 120 TVQATVHKLV DSGQLEFING GWCMHDEAVV HYIDMIDQTT LGHRVIKKQF KKIPRAGWQI 180 DPFGHSAVQA YLLGAELGFD SVHFARIDYQ DRATRKANKG LEVIWRGSRP VGSSSQIFTN 240 VFPVHYSAPV GFSFEVLAEN VIPVQDDMSL FDYNVQERVD DFVAAAIAQA NVTRTNHIMW 300 TMGDDFNYQY AESWFRNMDK LIQYVNKDGR VHALYSTPSI YTDAKHASNE SWPVKYDDYF 360 PYADSTNAYW TGYFTSRPTF KRYIRVYSGY YLAARQIEFL MGRSSLGLFT SSLEDAMGIA 420 QHHDAISGTA KQHTTDDYSK RIAIGASKVE KGVNAALTCL TNSNKTCVSS VPKFSQCPLL 480 NISYCPSTEE AISATKHLVV VVYNPLGWKR SDFIRVPVND EDLVVKSSDG NTVVSQIVVV 540 DNVTNNLRKL YVKAYLGVTA NKAPKYWLTF QASVPPMGWN SYFILKSTGS GYNNTEHVPV 600 VVPPSNNTIE AGQGHLKMSF SSASGQLERI FNSASGGDLP IQQSFLWYRS SKGDALDPQA 660 SGAYIFRPDG NTPTIASSSV RP |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
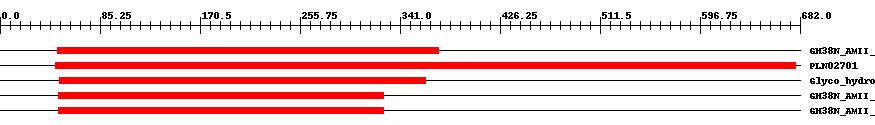 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd10809 | GH38N_AMII_GMII_SfManIII_like | 3.0e-77 | 49 | 374 | 352 | + N-terminal catalytic domain of Golgi alpha-mannosidase II, Spodoptera frugiperda Sf9 alpha-mannosidase III, and similar proteins; glycoside hydrolase family 38 (GH38). This subfamily is represented by Golgi alpha-mannosidase II (GMII, also known as mannosyl-oligosaccharide 1,3- 1,6-alpha mannosidase, EC 3.2.1.114, Man2A1), a monomeric, membrane-anchored class II alpha-mannosidase existing in the Golgi apparatus of eukaryotes. GMII plays a key role in the N-glycosylation pathway. It catalyzes the hydrolysis of the terminal both alpha-1,3-linked and alpha-1,6-linked mannoses from the high-mannose oligosaccharide GlcNAc(Man)5(GlcNAc)2 to yield GlcNAc(Man)3(GlcNAc)2(GlcNAc, N-acetylglucosmine), which is the committed step of complex N-glycan synthesis. GMII is activated by zinc or cobalt ions and is strongly inhibited by swainsonine. Inhibition of GMII provides a route to block cancer-induced changes in cell surface oligosaccharide structures. GMII has a pH optimum of 5.5-6.0, which is intermediate between those of acidic (lysosomal alpha-mannosidase) and neutral (ER/cytosolic alpha-mannosidase) enzymes. GMII is a retaining glycosyl hydrolase of family GH38 that employs a two-step mechanism involving the formation of a covalent glycosyl enzyme complex; two carboxylic acids positioned within the active site act in concert: one as a catalytic nucleophile and the other as a general acid/base catalyst. This subfamily also includes human alpha-mannosidase 2x (MX, also known as mannosyl-oligosaccharide 1,3- 1,6-alpha mannosidase, EC 3.2.1.114, Man2A2). MX is enzymatically and functionally very similar to GMII, and is thought to also function in the N-glycosylation pathway. Also found in this subfamily is class II alpha-mannosidase encoded by Spodoptera frugiperda Sf9 cell. This alpha-mannosidase is an integral membrane glycoprotein localized in the Golgi apparatus. It shows high sequence homology with mammalian Golgi alpha-mannosidase II(GMII). It can hydrolyze p-nitrophenyl alpha-D-mannopyranoside (pNP-alpha-Man), and it is inhibited by swainsonine. However, the Sf9 enzyme is stimulated by cobalt and can hydrolyze (Man)5(GlcNAc)2 to (Man)3(GlcNAc)2, but it cannot hydrolyze GlcNAc(Man)5(GlcNAc)2, which is distinct from that of GMII. Thus, this enzyme has been designated as Sf9 alpha-mannosidase III (SfManIII). It probably functions in an alternate N-glycan processing pathway in Sf9 cells. | ||
| PLN02701 | PLN02701 | 4.0e-84 | 47 | 678 | 702 | + alpha-mannosidase | ||
| pfam01074 | Glyco_hydro_38 | 3.0e-94 | 51 | 363 | 317 | + Glycosyl hydrolases family 38 N-terminal domain. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. | ||
| cd00451 | GH38N_AMII_euk | 5.0e-111 | 50 | 327 | 278 | + N-terminal catalytic domain of eukaryotic class II alpha-mannosidases; glycoside hydrolase family 38 (GH38). The family corresponds to a group of eukaryotic class II alpha-mannosidases (AlphaMII), which contain Golgi alpha-mannosidases II (GMII), the major broad specificity lysosomal alpha-mannosidases (LAM, MAN2B1), the noval core-specific lysosomal alpha 1,6-mannosidases (Epman, MAN2B2), and similar proteins. GMII catalyzes the hydrolysis of the terminal both alpha-1,3-linked and alpha-1,6-linked mannoses from the high-mannose oligosaccharide GlcNAc(Man)5(GlcNAc)2 to yield GlcNAc(Man)3(GlcNAc)2 (GlcNAc, N-acetylglucosmine), which is the committed step of complex N-glycan synthesis. LAM is a broad specificity exoglycosidase hydrolyzing all known alpha 1,2-, alpha 1,3-, and alpha 1,6-mannosidic linkages from numerous high mannose type oligosaccharides. Different from LAM, Epman can efficiently cleave only the alpha 1,6-linked mannose residue from (Man)3GlcNAc, but not (Man)3(GlcNAc)2 or other larger high mannose oligosaccharides, in the core of N-linked glycans. Members in this family are retaining glycosyl hydrolases of family GH38 that employs a two-step mechanism involving the formation of a covalent glycosyl enzyme complex. Two carboxylic acids positioned within the active site act in concert: one as a catalytic nucleophile and the other as a general acid/base catalyst. | ||
| cd10810 | GH38N_AMII_LAM_like | 9.0e-172 | 50 | 327 | 280 | + N-terminal catalytic domain of lysosomal alpha-mannosidase and similar proteins; glycoside hydrolase family 38 (GH38). The subfamily is represented by lysosomal alpha-mannosidase (LAM, Man2B1, EC 3.2.1.114), which is a broad specificity exoglycosidase hydrolyzing all known alpha 1,2-, alpha 1,3-, and alpha 1,6-mannosidic linkages from numerous high mannose type oligosaccharides. LAM is expressed in all tissues and in many species. In mammals, the absence of LAM can cause the autosomal recessive disease alpha-mannosidosis. LAM has an acidic pH optimum at 4.0-4.5. It is stimulated by zinc ion and is inhibited by cobalt ion and plant alkaloids, such as swainsonine (SW). LAM catalyzes hydrolysis by a double displacement mechanism in which a glycosyl-enzyme intermediate is formed and hydrolyzed via oxacarbenium ion-like transition states. A carboxylic acid in the active site acts as the catalytic nucleophile in the formation of the covalent intermediate while a second carboxylic acid acts as a general acid catalyst. The same residue is thought to assist in the hydrolysis (deglycosylation) step, this time acting as a general base. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0004559 | alpha-mannosidase activity |
| GO:0005975 | carbohydrate metabolic process |
| GO:0006013 | mannose metabolic process |
| GO:0008270 | zinc ion binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAL73068.1 | 0 | 45 | 639 | 33 | 635 | AC090873_14 Putative alpha-mannosidase [Oryza sativa] |
| GenBank | EEC66555.1 | 0 | 45 | 677 | 33 | 662 | hypothetical protein OsI_32713 [Oryza sativa Indica Group] |
| RefSeq | NP_001064140.1 | 0 | 45 | 677 | 33 | 662 | Os10g0140200 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | XP_002464752.1 | 0 | 44 | 680 | 34 | 670 | hypothetical protein SORBIDRAFT_01g026390 [Sorghum bicolor] |
| RefSeq | XP_002464754.1 | 0 | 144 | 680 | 1 | 537 | hypothetical protein SORBIDRAFT_01g026405 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1o7d_A | 0 | 50 | 333 | 14 | 298 | C Chain C, The Structure Of The Bovine Lysosomal A-Mannosidase Suggests A Novel Mechanism For Low Ph Activation |
| PDB | 1hxk_A | 0 | 48 | 670 | 49 | 703 | A Chain A, Golgi Alpha-Mannosidase Ii In Complex With Deoxymannojirimicin |
| PDB | 1hww_A | 0 | 48 | 670 | 49 | 703 | A Chain A, Golgi Alpha-Mannosidase Ii In Complex With Deoxymannojirimicin |
| PDB | 1hty_A | 0 | 48 | 670 | 49 | 703 | A Chain A, Golgi Alpha-Mannosidase Ii |
| PDB | 1ps3_A | 0 | 48 | 670 | 79 | 733 | A Chain A, Golgi Alpha-mannosidase Ii In Complex With Kifunensine |