

| Basic Information | |
|---|---|
| Species | Linum usitatissimum |
| Cazyme ID | Lus10041639 |
| Family | GH18 |
| Protein Properties | Length: 786 Molecular Weight: 86265.1 Isoelectric Point: 7.0431 |
| Chromosome | Chromosome/Scaffold: 272 Start: 1065594 End: 1069433 |
| Description | S-locus lectin protein kinase family protein |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH18 | 48 | 367 | 0 |
| AAAINATLYTHLICGYATINRTTYQVSISPSDRPHFANFTSAVRRHNPSVTTLYSISDGGDSNNSAAFSAMLNNSSHRKAFIQTAIQTARTHGFRGIDFS WSNDSPDDLANIGTLFREWRVQINSAAGERRLLLTAAFDYAPGMGVRNFPVGSIRNNLDWVHVMAQNLESSTAMEPDSPLDDPEFLSATRSGIDAWISVG KVPPEKLVLSLPLYGFAWKLANPAAAKGNYIGARVSGPAVNKEGGGFLSYGEIKEITNGSEILYNSTYAVNYCTVNGSVSVIFDGPEAIGAKVSYTTGMH LGGYYVWQIPFDYNGELALV | |||
| Full Sequence |
|---|
| Protein Sequence Length: 786 Download |
| MMPTVLLPSP NLVIILIIAS SLLSMANARH LIRGGYWVVS DFGPPGVAAA INATLYTHLI 60 CGYATINRTT YQVSISPSDR PHFANFTSAV RRHNPSVTTL YSISDGGDSN NSAAFSAMLN 120 NSSHRKAFIQ TAIQTARTHG FRGIDFSWSN DSPDDLANIG TLFREWRVQI NSAAGERRLL 180 LTAAFDYAPG MGVRNFPVGS IRNNLDWVHV MAQNLESSTA MEPDSPLDDP EFLSATRSGI 240 DAWISVGKVP PEKLVLSLPL YGFAWKLANP AAAKGNYIGA RVSGPAVNKE GGGFLSYGEI 300 KEITNGSEIL YNSTYAVNYC TVNGSVSVIF DGPEAIGAKV SYTTGMHLGG YYVWQIPFDY 360 NGELALVAAK EQGKKQSQLK KRERVLAGTV SVAVFLTIIL SIAAFYWIRI RGLGRFKALL 420 SFGGSVSNLR VEDIEGSGDF SRNDAVPNLR VYSFDEINKA TNGFSLQNKL GRGGYGPVYK 480 GVLPDGQDIA VKKLSETSTQ GLQEFKNEVQ LNARIQHVNL VRVLGYCIDN DEKLLIYEFM 540 PNNSLDKYLY DSAKRDILDW SRRVEIIQGI TQGLVYLQEY SRLPIIHRDL KASNILLDSD 600 FLPKISDFGI AKVFSKNGEE TNTVRIVGTI GYTPPEYFRQ GTYSVKSDVY GFGVLLLQII 660 TSKKVARMYG LAGDSSLQEY AFEVWEDGRG MEIVDSSLDD SSSSCKLLTC LRIALLCVQD 720 DPIDRPSMLQ VASMLRNETV VGSLFPKKPT FSFKRQGDNN VELPAVLQPA FCSIADGTFT 780 ELEGR* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
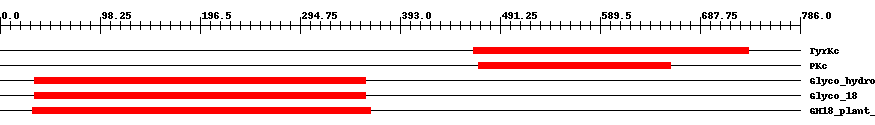 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| smart00219 | TyrKc | 2.0e-48 | 465 | 735 | 282 | + Tyrosine kinase, catalytic domain. Phosphotransferases. Tyrosine-specific kinase subfamily. | ||
| cd00180 | PKc | 4.0e-50 | 470 | 659 | 194 | + Catalytic domain of Protein Kinases. Protein Kinases (PKs), catalytic (c) domain. PKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine or tyrosine residues on protein substrates. The PK family is part of a larger superfamily that includes the catalytic domains of RIO kinases, aminoglycoside phosphotransferase, choline kinase, phosphoinositide 3-kinase (PI3K), and actin-fragmin kinase. PKs make up a large family of serine/threonine kinases, protein tyrosine kinases (PTKs), and dual-specificity PKs that phosphorylate both serine/threonine and tyrosine residues of target proteins. Majority of protein phosphorylation, about 95%, occurs on serine residues while only 1% occurs on tyrosine residues. Protein phosphorylation is a mechanism by which a wide variety of cellular proteins, such as enzymes and membrane channels, are reversibly regulated in response to certain stimuli. PKs often function as components of signal transduction pathways in which one kinase activates a second kinase, which in turn, may act on other kinases; this sequential action transmits a signal from the cell surface to target proteins, which results in cellular responses. The PK family is one of the largest known protein families with more than 100 homologous yeast enzymes and 550 human proteins. A fraction of PK family members are pseudokinases that lack crucial residues for catalytic activity. The mutiplicity of kinases allows for specific regulation according to substrate, tissue distribution, and cellular localization. PKs regulate many cellular processes including proliferation, division, differentiation, motility, survival, metabolism, cell-cycle progression, cytoskeletal rearrangement, immunity, and neuronal functions. Many kinases are implicated in the development of various human diseases including different types of cancer. | ||
| pfam00704 | Glyco_hydro_18 | 3.0e-51 | 34 | 359 | 337 | + Glycosyl hydrolases family 18. | ||
| smart00636 | Glyco_18 | 1.0e-53 | 34 | 359 | 346 | + Glyco_18 domain. | ||
| cd02879 | GH18_plant_chitinase_class_V | 7.0e-80 | 32 | 364 | 344 | + The class V plant chitinases have a glycosyl hydrolase family 18 (GH18) domain, but lack the chitin-binding domain present in other GH18 enzymes. The GH18 domain of the class V chitinases has endochitinase activity in some cases and no catalytic activity in others. Included in this family is a lectin found in black locust (Robinia pseudoacacia) bark, which binds chitin but lacks chitinase activity. Also included is a chitinase-related receptor-like kinase (CHRK1) from tobacco (Nicotiana tabacum), with an N-terminal GH18 domain and a C-terminal kinase domain, which is thought to be part of a plant signaling pathway. The GH18 domain of CHRK1 is expressed extracellularly where it binds chitin but lacks chitinase activity. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0004672 | protein kinase activity |
| GO:0005524 | ATP binding |
| GO:0005975 | carbohydrate metabolic process |
| GO:0006468 | protein phosphorylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CBI35714.1 | 0 | 3 | 755 | 392 | 1101 | unnamed protein product [Vitis vinifera] |
| EMBL | CBI35714.1 | 0 | 32 | 478 | 26 | 393 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_002263709.1 | 0 | 32 | 755 | 26 | 735 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002263792.1 | 0 | 7 | 755 | 1 | 724 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002324558.1 | 0 | 12 | 785 | 8 | 763 | predicted protein [Populus trichocarpa] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3alg_A | 0 | 32 | 380 | 4 | 353 | A Chain A, Crystal Structure Of Class V Chitinase (E115q Mutant) From Nicotiana Tobaccum In Complex With Nag4 |
| PDB | 3alf_A | 0 | 32 | 380 | 4 | 353 | A Chain A, Crystal Structure Of Class V Chitinase From Nicotiana Tobaccum |
| PDB | 3ulz_A | 0 | 448 | 738 | 16 | 309 | A Chain A, Crystal Structure Of Class V Chitinase From Nicotiana Tobaccum |
| PDB | 3uim_A | 0 | 448 | 738 | 16 | 309 | A Chain A, Structural Basis For The Impact Of Phosphorylation On Plant Receptor- Like Kinase Bak1 Activation |
| PDB | 3tl8_H | 0 | 448 | 738 | 24 | 317 | B Chain B, The Avrptob-Bak1 Complex Reveals Two Structurally Similar Kinaseinteracting Domains In A Single Type Iii Effector |