

| Basic Information | |
|---|---|
| Species | Malus domestica |
| Cazyme ID | MDP0000464809 |
| Family | CE10 |
| Protein Properties | Length: 952 Molecular Weight: 105549 Isoelectric Point: 6.4298 |
| Chromosome | Chromosome/Scaffold: 003994328 Start: 7360 End: 11712 |
| Description | alpha/beta-Hydrolases superfamily protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CE10 | 44 | 399 | 0 |
| EEPVVPANPTFQNGVATKNIHIDPNSSLSLRIFLPDTVLPLKAPNPTSRVGALLSPSPXCSNSDDGVVYRGYSPDQLVGRRHRKVPIFLQFHGGGFVSGS NDTSWNDAFCRRMAKLCDAIVVAVGYRLAPESPYPAAFEDGVTVLKWVAKQANLALVQKGRSRIFDSFGSSMVEPWLAAHGDPSRCVLLGVSCGANLADY VARKAVEAGDLLDPIKVVAQVLMYPFFIGSTPTRSEIKLANSYLFDKATCMLAWKLFQTEEEFDLDHPAGNPLMPAGRGPPLKTMPPTLTVVAQHDWMRD RGIAYSEELRKANVDAPLLDYKDTVHEFATLDVLLETPQAKACAEDITIWYKKEAS | |||
| Full Sequence |
|---|
| Protein Sequence Length: 952 Download |
| MPPLSLKLHS LFFKYHLRHQ LHNLTQSAQL QNTDPKFGIT SRPEEPVVPA NPTFQNGVAT 60 KNIHIDPNSS LSLRIFLPDT VLPLKAPNPT SRVGALLSPS PXCSNSDDGV VYRGYSPDQL 120 VGRRHRKVPI FLQFHGGGFV SGSNDTSWND AFCRRMAKLC DAIVVAVGYR LAPESPYPAA 180 FEDGVTVLKW VAKQANLALV QKGRSRIFDS FGSSMVEPWL AAHGDPSRCV LLGVSCGANL 240 ADYVARKAVE AGDLLDPIKV VAQVLMYPFF IGSTPTRSEI KLANSYLFDK ATCMLAWKLF 300 QTEEEFDLDH PAGNPLMPAG RGPPLKTMPP TLTVVAQHDW MRDRGIAYSE ELRKANVDAP 360 LLDYKDTVHE FATLDVLLET PQAKACAEDI TIWYKKEAST YTSHISHIVY ISPQGDSIGQ 420 ASNTLESYTM ASDLYVQPEF CGEITSNEVI GFDLNLSSMS CLPHPSLSAL EDDSANWVSP 480 FVDKTRNHKR LKQEHDANYG FKVGTNYSSF YTGSDSNMCT SGFTSLPTIQ FRDHISAHKR 540 RYLAAEAMEE AFATLMRDKE DESEEGGGSE DEIKLVQQLI ACAEAVACRD KAHASALLFE 600 LRANAQAFGT SFQRVSSCFV QGLAXRLALV QPLGAGGVID PNVKSKPFSA EKGPFSAEKD 660 EALHLVYEIC PQIQFGHFVA NASILEAFEG GSSVHVIDLG MTLGLQHGYQ WRDLIDRLAN 720 RPGQPLHXLR ITGVGSSSER LQAIGDDLKL HAQSMKINXD FSEVESNLEN LKPQDFHLVD 780 GEILIINSIL QLHCLVKESR GTLNSALQTL HELSPKLMVL VEQDTSHNGP FFLGRFMEAL 840 HNYSAIFDSL DAMLPKYDTR RVKMEQFYFG EEIKNIVSCE GPARVERHER VDQWRRRMRR 900 AGFQPAPLRT MAQAVKWLET NACEGYTVVE DKGCLVLGWK SKPIIATSCW K* 960 |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd00312 | Esterase_lipase | 4.0e-7 | 115 | 238 | 133 | + Esterases and lipases (includes fungal lipases, cholinesterases, etc.) These enzymes act on carboxylic esters (EC: 3.1.1.-). The catalytic apparatus involves three residues (catalytic triad): a serine, a glutamate or aspartate and a histidine.These catalytic residues are responsible for the nucleophilic attack on the carbonyl carbon atom of the ester bond. In contrast with other alpha/beta hydrolase fold family members, p-nitrobenzyl esterase and acetylcholine esterase have a Glu instead of Asp at the active site carboxylate. | ||
| PRK10162 | PRK10162 | 5.0e-9 | 86 | 196 | 111 | + acetyl esterase; Provisional | ||
| COG0657 | Aes | 3.0e-39 | 106 | 390 | 286 | + Esterase/lipase [Lipid metabolism] | ||
| pfam07859 | Abhydrolase_3 | 1.0e-62 | 131 | 372 | 242 | + alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. | ||
| pfam03514 | GRAS | 8.0e-122 | 575 | 951 | 385 | + GRAS family transcription factor. Sequence analysis of the products of the GRAS (GAI, RGA, SCR) gene family indicates that they share a variable amino-terminus and a highly conserved carboxyl-terminus that contains five recognisable motifs. Proteins in the GRAS family are transcription factors that seem to be involved in development and other processes. Mutation of the SCARECROW (SCR) gene results in a radial pattern defect, loss of a ground tissue layer, in the root. The PAT1 protein is involved in phytochrome A signal transduction. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0008152 | metabolic process |
| GO:0016787 | hydrolase activity |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| DDBJ | BAF37945.1 | 0 | 1 | 407 | 1 | 407 | hypothetical protein [Malus x domestica] |
| RefSeq | XP_002263040.1 | 0 | 430 | 951 | 1 | 518 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002318703.1 | 0 | 435 | 951 | 5 | 515 | GRAS family transcription factor [Populus trichocarpa] |
| RefSeq | XP_002322227.1 | 0 | 435 | 951 | 5 | 516 | GRAS family transcription factor [Populus trichocarpa] |
| RefSeq | XP_002515010.1 | 0 | 430 | 951 | 1 | 505 | DELLA protein GAI1, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2zsi_A | 2e-34 | 128 | 374 | 113 | 331 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2zsh_A | 2e-34 | 128 | 374 | 113 | 331 | B Chain B, Structural Basis Of Gibberellin(Ga3)-Induced Della Recognition By The Gibberellin Receptor |
| PDB | 3ed1_F | 3e-30 | 129 | 354 | 113 | 310 | B Chain B, Structural Basis Of Gibberellin(Ga3)-Induced Della Recognition By The Gibberellin Receptor |
| PDB | 3ed1_E | 3e-30 | 129 | 354 | 113 | 310 | B Chain B, Structural Basis Of Gibberellin(Ga3)-Induced Della Recognition By The Gibberellin Receptor |
| PDB | 3ed1_D | 3e-30 | 129 | 354 | 113 | 310 | B Chain B, Structural Basis Of Gibberellin(Ga3)-Induced Della Recognition By The Gibberellin Receptor |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
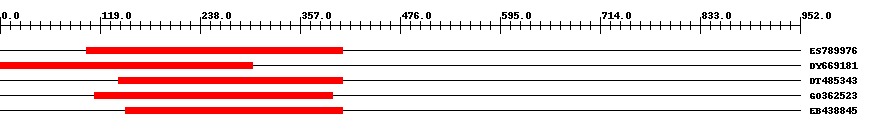 | ||||
| Hit | Length | Start | End | EValue |
| ES789976 | 333 | 103 | 407 | 0 |
| DY669181 | 307 | 1 | 300 | 0 |
| DT485343 | 269 | 141 | 407 | 0 |
| GO362523 | 304 | 113 | 396 | 0 |
| EB438845 | 276 | 149 | 407 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |