

| Basic Information | |
|---|---|
| Species | Medicago truncatula |
| Cazyme ID | Medtr4g128720.1 |
| Family | GT1 |
| Protein Properties | Length: 468 Molecular Weight: 51794.9 Isoelectric Point: 6.7154 |
| Chromosome | Chromosome/Scaffold: 4 Start: 45048980 End: 45051028 |
| Description | UDP-glucosyl transferase 78D2 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT1 | 287 | 441 | 1.1e-30 |
| SVVYISFGSRIVPPPHELNALAECLEECGCPFIWVFKGNPEETLPNGFTERTKTKGKFVAWAPQMEILKHSAVGMCLTHSGWNSVLDCIVGGVPMVSRPF FGDQRLNARMLESIWGIGVGVDNGVLTKESTMKALNLIMSTEEGKIMREKILKLK | |||
| Full Sequence |
|---|
| Protein Sequence Length: 468 Download |
| MFSIARLKNW MSRDKGESVN VEKLHVAVLA FPFGTHAGPL LNLARRIAVD AHKVTFSFLC 60 TSRTNAALFS GSKDYDEFLP NIKHYDVHDG LPEGYVPSGH PLEPIFIFIK AMPDNYKSVM 120 VKAVAETGKN ITCLVTDAFY WFGADLAKEM HAKWVPLWTA GPHSLLTHVF TDLIREKIGS 180 KEVDDTKTVD FLPGFPELHV SDFPEGVIGD IDGPFSTMLH KMGLELPRAT AVAINSFSTV 240 HPLIENELNS QFKLLLNVGP FILTTPQRMV SDEHGCLAWL NQCEKFSVVY ISFGSRIVPP 300 PHELNALAEC LEECGCPFIW VFKGNPEETL PNGFTERTKT KGKFVAWAPQ MEILKHSAVG 360 MCLTHSGWNS VLDCIVGGVP MVSRPFFGDQ RLNARMLESI WGIGVGVDNG VLTKESTMKA 420 LNLIMSTEEG KIMREKILKL KESALKAVER NGTSANNFDT LIKIVTS* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02863 | PLN02863 | 9.0e-38 | 23 | 467 | 488 | + UDP-glucoronosyl/UDP-glucosyl transferase family protein | ||
| PLN03007 | PLN03007 | 5.0e-38 | 19 | 467 | 489 | + UDP-glucosyltransferase family protein | ||
| PLN02173 | PLN02173 | 2.0e-38 | 21 | 467 | 477 | + UDP-glucosyl transferase family protein | ||
| PLN02555 | PLN02555 | 2.0e-39 | 25 | 458 | 485 | + limonoid glucosyltransferase | ||
| PLN02410 | PLN02410 | 4.0e-52 | 26 | 462 | 450 | + UDP-glucoronosyl/UDP-glucosyl transferase family protein | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0008152 | metabolic process |
| GO:0016758 | transferase activity, transferring hexosyl groups |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| Swiss-Prot | A6XNC6 | 0 | 11 | 467 | 1 | 454 | UGFGT_MEDTR RecName: Full=Flavonoid 3-O-glucosyltransferase; AltName: Full=UDP glucose:flavonoid 3-O-glucosyltransferase; AltName: Full=UDP-glycosyltransferase 78G1 |
| GenBank | ACH56523.2 | 0 | 25 | 465 | 10 | 444 | flavonoid 3-glucosyltransferase [Gossypium hirsutum] |
| DDBJ | BAE72453.1 | 0 | 25 | 467 | 16 | 455 | UDP-glucose: flavonol 3-O-glucosyltransferase [Rosa hybrid cultivar] |
| RefSeq | NP_197207.1 | 0 | 25 | 467 | 12 | 458 | UGT78D2 (UDP-GLUCOSYL TRANSFERASE 78D2); UDP-glycosyltransferase/ anthocyanidin 3-O-glucosyltransferase/ quercetin 3-O-glucosyltransferase/ transferase, transferring glycosyl groups [Arabidopsis thaliana] |
| RefSeq | XP_002533888.1 | 0 | 25 | 466 | 13 | 453 | UDP-glucosyltransferase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3hbj_A | 0 | 11 | 467 | 1 | 454 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 3hbf_A | 0 | 11 | 467 | 1 | 454 | A Chain A, Structure Of Ugt78g1 Complexed With Myricetin And Udp |
| PDB | 2c9z_A | 0 | 25 | 466 | 9 | 451 | A Chain A, Structure Of Ugt78g1 Complexed With Myricetin And Udp |
| PDB | 2c1z_A | 0 | 25 | 466 | 9 | 451 | A Chain A, Structure Of Ugt78g1 Complexed With Myricetin And Udp |
| PDB | 2c1x_A | 0 | 25 | 466 | 9 | 451 | A Chain A, Structure And Activity Of A Flavonoid 3-O Glucosyltransferase Reveals The Basis For Plant Natural Product Modification |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
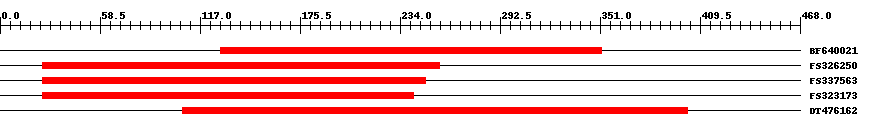 | ||||
| Hit | Length | Start | End | EValue |
| BF640021 | 224 | 129 | 352 | 0 |
| FS326250 | 233 | 25 | 257 | 0 |
| FS337563 | 225 | 25 | 249 | 0 |
| FS323173 | 218 | 25 | 242 | 0 |
| DT476162 | 299 | 107 | 402 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |