

| Basic Information | |
|---|---|
| Species | Manihot esculenta |
| Cazyme ID | cassava4.1_023739m |
| Family | GH1 |
| Protein Properties | Length: 310 Molecular Weight: 35126.3 Isoelectric Point: 9.0444 |
| Chromosome | Chromosome/Scaffold: 00109 Start: 142424 End: 144625 |
| Description | beta glucosidase 40 |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH1 | 23 | 184 | 1.19951e-42 |
| AETCFEKFGDSVKHWITLNEPRTFTIQGYDVCLQAPGRCSIVLRLLCRAGNSATEPYIVAHNFLLSHGTVADIYKKKYKSLQAKQGGSLGISFDVMWFEP ASNSTEDIEATQRAQDFQLGWFIEPLLFGDYPSSMKTRVGNRLPLFSKSEAALIKGSLDFPM | |||
| Full Sequence |
|---|
| Protein Sequence Length: 310 Download |
| GTRAINLAGV DHYNKLINAL IAAETCFEKF GDSVKHWITL NEPRTFTIQG YDVCLQAPGR 60 CSIVLRLLCR AGNSATEPYI VAHNFLLSHG TVADIYKKKY KSLQAKQGGS LGISFDVMWF 120 EPASNSTEDI EATQRAQDFQ LGWFIEPLLF GDYPSSMKTR VGNRLPLFSK SEAALIKGSL 180 DFPMQNLLFF TFLINSLFLL LPTFRGLKPI GDRIFAGMND SNTFTSIKDA LKDEKRIKYH 240 NDYLTNLLAS IKEDGCNVKG YFVWSLLDNW EWAAGYSSRF GLFFVDYKDK LKRYPKDSVK 300 WFKNFLTST* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG2723 | BglB | 4.0e-51 | 1 | 308 | 384 | + Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism] | ||
| TIGR03356 | BGL | 5.0e-58 | 1 | 302 | 373 | + beta-galactosidase. | ||
| PLN02998 | PLN02998 | 9.0e-63 | 23 | 309 | 330 | + beta-glucosidase | ||
| PLN02849 | PLN02849 | 5.0e-65 | 11 | 306 | 337 | + beta-glucosidase | ||
| PLN02814 | PLN02814 | 7.0e-66 | 11 | 309 | 341 | + beta-glucosidase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ADD17684.1 | 0 | 1 | 308 | 115 | 504 | beta-glucosidase [Vitis vinifera] |
| EMBL | CBI27264.1 | 0 | 1 | 308 | 175 | 564 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_002274662.1 | 0 | 1 | 308 | 115 | 504 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002311330.1 | 0 | 1 | 309 | 119 | 512 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002512097.1 | 0 | 1 | 309 | 115 | 504 | beta-glucosidase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3gnr_A | 0 | 1 | 308 | 100 | 488 | A Chain A, Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum |
| PDB | 3gnp_A | 0 | 1 | 308 | 100 | 488 | A Chain A, Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum |
| PDB | 3gno_A | 0 | 1 | 308 | 100 | 488 | A Chain A, Crystal Structure Of A Rice Os3bglu6 Beta-glucosidase |
| PDB | 3ptq_B | 0 | 11 | 306 | 161 | 503 | A Chain A, Crystal Structure Of A Rice Os3bglu6 Beta-glucosidase |
| PDB | 3ptq_A | 0 | 11 | 306 | 161 | 503 | A Chain A, Crystal Structure Of A Rice Os3bglu6 Beta-glucosidase |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| coumarin biosynthesis (via 2-coumarate) | RXN-8036 | EC-3.2.1.21 | β-glucosidase |
| linamarin degradation | RXN-5341 | EC-3.2.1.21 | β-glucosidase |
| linustatin bioactivation | RXN-13602 | EC-3.2.1.21 | β-glucosidase |
| linustatin bioactivation | RXN-5341 | EC-3.2.1.21 | β-glucosidase |
| lotaustralin degradation | RXN-9674 | EC-3.2.1.21 | β-glucosidase |
| neolinustatin bioactivation | RXN-13603 | EC-3.2.1.21 | β-glucosidase |
| neolinustatin bioactivation | RXN-9674 | EC-3.2.1.21 | β-glucosidase |
| taxiphyllin bioactivation | RXN-13600 | EC-3.2.1.21 | β-glucosidase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
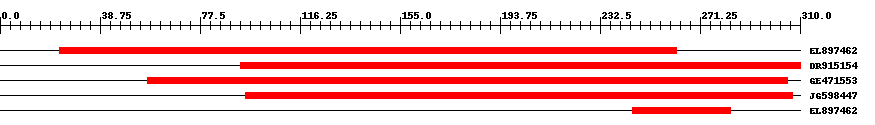 | ||||
| Hit | Length | Start | End | EValue |
| EL897462 | 288 | 23 | 262 | 0 |
| DR915154 | 267 | 93 | 310 | 0 |
| GE471553 | 287 | 57 | 305 | 0 |
| JG598447 | 261 | 95 | 307 | 0 |
| EL897462 | 39 | 245 | 283 | 0.000001 |
| Orthologous Group | |||||
|---|---|---|---|---|---|
| Species | ID | ||||
| Arabidopsis lyrata | 918586 | ||||
| Carica papaya | evm.model.supercontig_80.123 | ||||
| Oryza sativa | LOC_Os06g46940.1 | ||||
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |