

| Basic Information | |
|---|---|
| Species | Carica papaya |
| Cazyme ID | evm.model.supercontig_106.45 |
| Family | CBM22 |
| Protein Properties | Length: 585 Molecular Weight: 64974 Isoelectric Point: 8.5966 |
| Chromosome | Chromosome/Scaffold: 106 Start: 572838 End: 575957 |
| Description | Glycosyl hydrolase family 10 protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM22 | 53 | 168 | 2.6e-22 |
| IITNPELNQGLKGWSTFGDAKIQHRVAGSNSFIVAHTRSQPHDSVSQTLYLQSNKLYTFSAWIRVSEGKTPVKAIFKTKSGYKYAGAVVAESNCWSMLKG GLTVDASGPAELYFET | |||
| GH10 | 230 | 491 | 0 |
| IGFPFGCAINRNILNNNAYQSWFSSRFTVTTFENEMKWASTEPSQGHEDYSTADAMVQFAKKNGIAIRGHNVFWDDPKYQSGWVSSLSPNDLNAAATKRI NSVMNRYKGQVIGWDVVNENLHFSFFESKLGANASAVFYGEAHKTDPSTTLFMNEYNTVEDSRDGQATPAKYLEKLRSIQSLPGNGNMGIGLESHFSSSP PNIPYMRSAIDTLAATGLPVWLTEVDVQSGGNQAQSLEQILREAHSHPKVRGIVIWSAWSPN | |||
| Full Sequence |
|---|
| Protein Sequence Length: 585 Download |
| MKLGEKNLQF YFLLVLPYAL LFPGLETNAL SYDYTASIQC LENPQKAQYG GGIITNPELN 60 QGLKGWSTFG DAKIQHRVAG SNSFIVAHTR SQPHDSVSQT LYLQSNKLYT FSAWIRVSEG 120 KTPVKAIFKT KSGYKYAGAV VAESNCWSML KGGLTVDASG PAELYFETDN TSVEIWIDSI 180 SLQPFTQQEW KSHQDQSIKK IRKKNVRIQA VDKLGNPLPN TTVSISPKKI GFPFGCAINR 240 NILNNNAYQS WFSSRFTVTT FENEMKWAST EPSQGHEDYS TADAMVQFAK KNGIAIRGHN 300 VFWDDPKYQS GWVSSLSPND LNAAATKRIN SVMNRYKGQV IGWDVVNENL HFSFFESKLG 360 ANASAVFYGE AHKTDPSTTL FMNEYNTVED SRDGQATPAK YLEKLRSIQS LPGNGNMGIG 420 LESHFSSSPP NIPYMRSAID TLAATGLPVW LTEVDVQSGG NQAQSLEQIL REAHSHPKVR 480 GIVIWSAWSP NGCYRMCLTD NNFHNLPTGD VVDKLLREWG GGATVKGKTD QNGFFQSSLF 540 HGDYEIKVQV NHPSKLPSSS SHHTFKLNST DDESKQTRLL LIKV* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam02018 | CBM_4_9 | 7.0e-5 | 60 | 172 | 123 | + Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. | ||
| COG3693 | XynA | 2.0e-37 | 228 | 485 | 285 | + Beta-1,4-xylanase [Carbohydrate transport and metabolism] | ||
| pfam00331 | Glyco_hydro_10 | 9.0e-58 | 232 | 485 | 271 | + Glycosyl hydrolase family 10. | ||
| smart00633 | Glyco_10 | 3.0e-82 | 265 | 512 | 270 | + Glycosyl hydrolase family 10. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAN10199.1 | 0 | 1 | 584 | 1 | 584 | endoxylanase [Carica papaya] |
| RefSeq | XP_002264556.1 | 0 | 23 | 581 | 103 | 654 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002264837.1 | 0 | 1 | 584 | 17 | 596 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002313180.1 | 0 | 38 | 562 | 1 | 524 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002532418.1 | 0 | 24 | 580 | 15 | 567 | Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1nq6_A | 4e-32 | 234 | 512 | 15 | 298 | A Chain A, Crystal Structure Of The Catalytic Domain Of Xylanase A From Streptomyces Halstedii Jm8 |
| PDB | 1xas_A | 4e-31 | 221 | 485 | 3 | 266 | A Chain A, Crystal Structure, At 2.6 Angstroms Resolution, Of The Streptomyces Lividans Xylanase A, A Member Of The F Family Of Beta-1,4-D-Glycanses |
| PDB | 1e0v_A | 4e-31 | 221 | 485 | 3 | 266 | A Chain A, Xylanase 10a From Sreptomyces Lividans. Cellobiosyl-Enzyme Intermediate At 1.7 A |
| PDB | 1v0n_A | 4e-31 | 221 | 485 | 3 | 266 | A Chain A, Xylanase 10a From Sreptomyces Lividans. Cellobiosyl-Enzyme Intermediate At 1.7 A |
| PDB | 1v0m_A | 4e-31 | 221 | 485 | 3 | 266 | A Chain A, Xylanase 10a From Sreptomyces Lividans. Cellobiosyl-Enzyme Intermediate At 1.7 A |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| (1,4)-β-xylan degradation | 3.2.1.8-RXN | EC-3.2.1.8 | endo-1,4-β-xylanase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
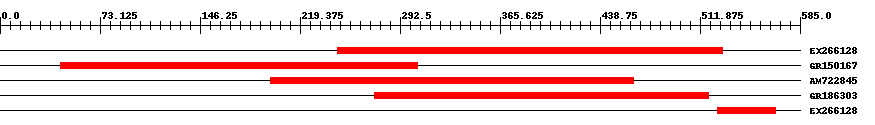 | ||||
| Hit | Length | Start | End | EValue |
| EX266128 | 282 | 247 | 528 | 0 |
| GR150167 | 263 | 44 | 305 | 0 |
| AM722845 | 268 | 198 | 463 | 0 |
| GR186303 | 245 | 274 | 518 | 0 |
| EX266128 | 43 | 525 | 567 | 0.00000005 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |