

| Basic Information | |
|---|---|
| Species | Carica papaya |
| Cazyme ID | evm.model.supercontig_196.11 |
| Family | GH77 |
| Protein Properties | Length: 302 Molecular Weight: 34393.5 Isoelectric Point: 7.1456 |
| Chromosome | Chromosome/Scaffold: 196 Start: 161746 End: 189091 |
| Description | disproportionating enzyme |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH77 | 1 | 302 | 0 |
| MQVLPLVPPDEGGSPYSGKDANCGNTLLISLEELVKDGLLSKDELPEPINAECVNFSIVSKLKDSLITKAAWRLILSEGELRSELHDFRGDPNISSWLED AAYFAAINNSLNTFSWYHWPEPLKNRHLAALEGIYDSQKDFIELFIAKQFLLQRQWQKVRKYAHLKGVSIMGDMPIYVDYHSADVWANKKQFLLNKKGFP LLVSGVPPDLFSKTGQLWGSPLYDWKAMEKDGFSWWVRRIQRAQDLYDEFRIDHFRGFTGFWAVPSEAKVAMVGRWKVGPGKALFDAISRAVGKIKIIAE DL | |||
| Full Sequence |
|---|
| Protein Sequence Length: 302 Download |
| MQVLPLVPPD EGGSPYSGKD ANCGNTLLIS LEELVKDGLL SKDELPEPIN AECVNFSIVS 60 KLKDSLITKA AWRLILSEGE LRSELHDFRG DPNISSWLED AAYFAAINNS LNTFSWYHWP 120 EPLKNRHLAA LEGIYDSQKD FIELFIAKQF LLQRQWQKVR KYAHLKGVSI MGDMPIYVDY 180 HSADVWANKK QFLLNKKGFP LLVSGVPPDL FSKTGQLWGS PLYDWKAMEK DGFSWWVRRI 240 QRAQDLYDEF RIDHFRGFTG FWAVPSEAKV AMVGRWKVGP GKALFDAISR AVGKIKIIAE 300 DL |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK14510 | PRK14510 | 1.0e-57 | 2 | 302 | 321 | + putative bifunctional 4-alpha-glucanotransferase/glycogen debranching enzyme; Provisional | ||
| COG1640 | MalQ | 2.0e-70 | 2 | 302 | 309 | + 4-alpha-glucanotransferase [Carbohydrate transport and metabolism] | ||
| pfam02446 | Glyco_hydro_77 | 6.0e-110 | 2 | 302 | 306 | + 4-alpha-glucanotransferase. These enzymes EC:2.4.1.25 transfer a segment of a (1,4)-alpha-D-glucan to a new 4-position in an acceptor, which may be glucose or (1,4)-alpha-D-glucan. | ||
| PRK14508 | PRK14508 | 2.0e-118 | 2 | 302 | 304 | + 4-alpha-glucanotransferase; Provisional | ||
| PLN02635 | PLN02635 | 0 | 2 | 302 | 305 | + disproportionating enzyme | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004134 | 4-alpha-glucanotransferase activity |
| GO:0005975 | carbohydrate metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1X1N | 0 | 2 | 302 | 66 | 370 | A Chain A, Structure Determination And Refinement At 1.8 A Resolution Of Disproportionating Enzyme From Potato |
| RefSeq | NP_201291.1 | 0 | 2 | 302 | 122 | 422 | DPE1 (DISPROPORTIONATING ENZYME); 4-alpha-glucanotransferase/ catalytic/ cation binding [Arabidopsis thaliana] |
| Swiss-Prot | Q06801 | 0 | 2 | 302 | 118 | 422 | DPEP_SOLTU RecName: Full=4-alpha-glucanotransferase, chloroplastic/amyloplastic; AltName: Full=Amylomaltase; AltName: Full=Disproportionating enzyme; Short=D-enzyme; Flags: Precursor |
| RefSeq | XP_002310024.1 | 0 | 2 | 302 | 116 | 420 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002528681.1 | 0 | 2 | 302 | 125 | 425 | 4-alpha-glucanotransferase, chloroplast precursor, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1x1n_A | 0 | 2 | 302 | 66 | 370 | A Chain A, Structure Determination And Refinement At 1.8 A Resolution Of Disproportionating Enzyme From Potato |
| PDB | 1tz7_B | 0 | 2 | 302 | 62 | 355 | A Chain A, Aquifex Aeolicus Amylomaltase |
| PDB | 1tz7_A | 0 | 2 | 302 | 62 | 355 | A Chain A, Aquifex Aeolicus Amylomaltase |
| PDB | 1esw_A | 0 | 2 | 302 | 45 | 342 | A Chain A, Aquifex Aeolicus Amylomaltase |
| PDB | 1cwy_A | 0 | 2 | 302 | 45 | 342 | A Chain A, Crystal Structure Of Amylomaltase From Thermus Aquaticus, A Glycosyltransferase Catalysing The Production Of Large Cyclic Glucans |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| starch degradation I | RXN-1828 | EC-2.4.1.25 | 4-α-glucanotransferase |
| starch degradation II | RXN-12391 | EC-2.4.1.25 | 4-α-glucanotransferase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
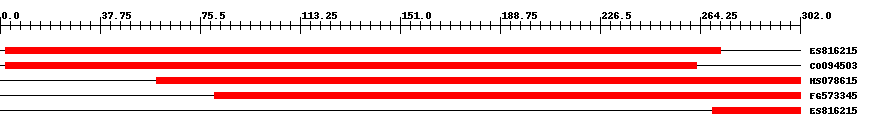 | ||||
| Hit | Length | Start | End | EValue |
| ES816215 | 271 | 2 | 272 | 0 |
| CO094503 | 262 | 2 | 263 | 0 |
| HS078615 | 244 | 59 | 302 | 0 |
| FG573345 | 222 | 81 | 302 | 0 |
| ES816215 | 34 | 269 | 302 | 0.000007 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |