

| Basic Information | |
|---|---|
| Species | Citrus sinensis |
| Cazyme ID | orange1.1g041546m |
| Family | AA7 |
| Protein Properties | Length: 491 Molecular Weight: 55268.7 Isoelectric Point: 6.8491 |
| Chromosome | Chromosome/Scaffold: 00008 Start: 2435458 End: 2437111 |
| Description | FAD-binding Berberine family protein |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA7 | 76 | 171 | 3.4e-32 |
| NLRFSTPNTPKPQVIITPLDVSQVQAAIKCSKKHGLQIRVRSGGHDFEGLSYVSHVPFVVIDLLNLSEISVDAAEQTAWVQAGATLGQLYYRIAEK | |||
| Full Sequence |
|---|
| Protein Sequence Length: 491 Download |
| MKSPFSPIFP FVFALLLSYH IRVTSADFHA TSAPVADLES LIQCLSMHSD NSSISKVIYT 60 QINSSYSSVL NFSIQNLRFS TPNTPKPQVI ITPLDVSQVQ AAIKCSKKHG LQIRVRSGGH 120 DFEGLSYVSH VPFVVIDLLN LSEISVDAAE QTAWVQAGAT LGQLYYRIAE KRYGAMLRKY 180 GLAADNIVDA RLTDANGRLL DRKSMGEDLF WAIQGGGIGA SFGVIVPWKV RLVIVPSTVT 240 RFRVSRSLEQ NATKIVHKWQ LLPLMQESFP ELGLTKEDCI EMSWIESAHD LAGFNKGDPL 300 DLLLDRNSRT NGVAEDAATN GFFKSKSDYV KQPIPENAFE GIYDNFYEED GETAFMLLVP 360 YGGKMSEISE SETPYPHRAG NIYQILYTVT WGEDETSQSH IDWIRRLYSH MTPYVSENPR 420 EAYINYRDLD IGTNNRGHTS IKQASIWGSK YFKNNFKRLV HVKTMVDPYD FFKNEQSIPP 480 LTSWRKKNGD * |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
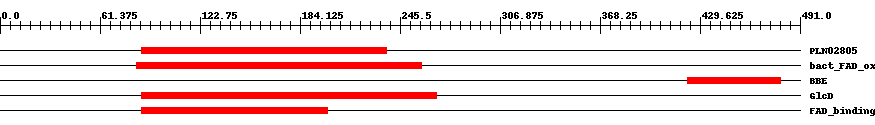 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02805 | PLN02805 | 0.003 | 87 | 237 | 182 | + D-lactate dehydrogenase [cytochrome] | ||
| TIGR01679 | bact_FAD_ox | 0.0004 | 84 | 259 | 201 | + FAD-linked oxidoreductase. This model represents a family of bacterial oxidoreductases with covalently linked FAD, closely related to two different eukaryotic oxidases, L-gulonolactone oxidase (EC 1.1.3.8) from rat and D-arabinono-1,4-lactone oxidase (EC 1.1.3.37) from Saccharomyces cerevisiae. | ||
| pfam08031 | BBE | 2.0e-15 | 422 | 479 | 58 | + Berberine and berberine like. This domain is found in the berberine bridge and berberine bridge- like enzymes which are involved in the biosynthesis of numerous isoquinoline alkaloids. They catalyze the transformation of the N-methyl group of (S)-reticuline into the C-8 berberine bridge carbon of (S)-scoulerine. | ||
| COG0277 | GlcD | 7.0e-20 | 87 | 268 | 213 | + FAD/FMN-containing dehydrogenases [Energy production and conversion] | ||
| pfam01565 | FAD_binding_4 | 1.0e-23 | 87 | 201 | 139 | + FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidises the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0008762 | UDP-N-acetylmuramate dehydrogenase activity |
| GO:0016491 | oxidoreductase activity |
| GO:0050660 | flavin adenine dinucleotide binding |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | XP_002330610.1 | 0 | 39 | 484 | 29 | 533 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002333020.1 | 0 | 1 | 484 | 1 | 533 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002333338.1 | 0 | 1 | 484 | 1 | 533 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002333339.1 | 0 | 1 | 484 | 1 | 533 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002334045.1 | 0 | 23 | 484 | 22 | 531 | predicted protein [Populus trichocarpa] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3vte_A | 0 | 39 | 481 | 5 | 513 | A Chain A, Crystal Structure Of Tetrahydrocannabinolic Acid Synthase From Cannabis Sativa |
| PDB | 4dns_B | 0 | 58 | 481 | 26 | 496 | A Chain A, Crystal Structure Of Bermuda Grass Isoallergen Bg60 Provides Insight Into The Various Cross-Allergenicity Of The Pollen Group 4 Allergens |
| PDB | 4dns_A | 0 | 58 | 481 | 26 | 496 | A Chain A, Crystal Structure Of Bermuda Grass Isoallergen Bg60 Provides Insight Into The Various Cross-Allergenicity Of The Pollen Group 4 Allergens |
| PDB | 3tsj_B | 0 | 34 | 481 | 4 | 496 | A Chain A, Crystal Structure Of Bermuda Grass Isoallergen Bg60 Provides Insight Into The Various Cross-Allergenicity Of The Pollen Group 4 Allergens |
| PDB | 3tsj_A | 0 | 34 | 481 | 4 | 496 | A Chain A, Crystal Structure Of Bermuda Grass Isoallergen Bg60 Provides Insight Into The Various Cross-Allergenicity Of The Pollen Group 4 Allergens |