You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000265_01356
You are here: Home > Sequence: MGYG000000265_01356
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides nordii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides nordii | |||||||||||
| CAZyme ID | MGYG000000265_01356 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 80917; End: 85233 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01095 | Pectinesterase | 2.24e-25 | 1022 | 1302 | 17 | 298 | Pectinesterase. |
| PLN02432 | PLN02432 | 1.86e-21 | 1031 | 1305 | 35 | 291 | putative pectinesterase |
| PLN02314 | PLN02314 | 7.29e-21 | 1022 | 1308 | 295 | 577 | pectinesterase |
| COG4677 | PemB | 5.16e-20 | 1033 | 1218 | 109 | 331 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02665 | PLN02665 | 1.57e-19 | 1061 | 1305 | 148 | 359 | pectinesterase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74167.1 | 0.0 | 1 | 1438 | 1 | 1435 |
| QCD38476.1 | 0.0 | 2 | 1438 | 3 | 1476 |
| QCP72166.1 | 0.0 | 2 | 1438 | 3 | 1476 |
| QCD41145.1 | 0.0 | 2 | 1438 | 3 | 1495 |
| QNT65238.1 | 1.06e-235 | 408 | 1438 | 88 | 1133 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 4.85e-19 | 1005 | 1244 | 26 | 307 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 2NSP_A | 1.37e-15 | 1010 | 1309 | 4 | 341 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1XG2_A | 2.78e-12 | 1018 | 1305 | 16 | 303 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 4PMH_A | 5.53e-10 | 1066 | 1224 | 132 | 298 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9LUL8 | 2.11e-18 | 1037 | 1307 | 689 | 959 | Putative pectinesterase/pectinesterase inhibitor 26 OS=Arabidopsis thaliana OX=3702 GN=PME26 PE=2 SV=1 |
| Q9FK05 | 2.36e-17 | 1020 | 1308 | 287 | 576 | Probable pectinesterase/pectinesterase inhibitor 61 OS=Arabidopsis thaliana OX=3702 GN=PME61 PE=2 SV=1 |
| Q43867 | 5.40e-17 | 1022 | 1308 | 296 | 578 | Pectinesterase 1 OS=Arabidopsis thaliana OX=3702 GN=PME1 PE=1 SV=1 |
| Q43111 | 4.91e-16 | 1033 | 1307 | 298 | 571 | Pectinesterase 3 OS=Phaseolus vulgaris OX=3885 GN=MPE3 PE=2 SV=1 |
| O22256 | 6.06e-16 | 1025 | 1305 | 265 | 546 | Probable pectinesterase/pectinesterase inhibitor 20 OS=Arabidopsis thaliana OX=3702 GN=PME20 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
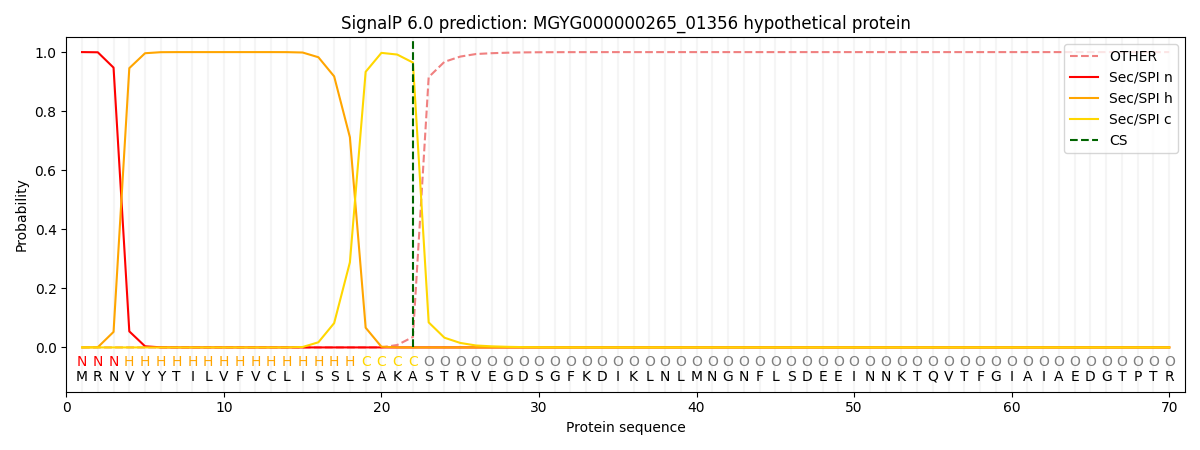
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000311 | 0.998971 | 0.000239 | 0.000157 | 0.000161 | 0.000153 |
