You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000289_01016
You are here: Home > Sequence: MGYG000000289_01016
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella buccae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella buccae | |||||||||||
| CAZyme ID | MGYG000000289_01016 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 51252; End: 54074 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 523 | 768 | 2.5e-40 | 0.8333333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.04e-16 | 476 | 722 | 51 | 331 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02432 | PLN02432 | 4.27e-12 | 521 | 728 | 30 | 220 | putative pectinesterase |
| PLN02708 | PLN02708 | 8.68e-10 | 521 | 735 | 260 | 484 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02488 | PLN02488 | 1.51e-09 | 506 | 722 | 199 | 417 | probable pectinesterase/pectinesterase inhibitor |
| PLN02745 | PLN02745 | 1.30e-08 | 526 | 735 | 309 | 518 | Putative pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH16322.1 | 1.38e-317 | 38 | 938 | 562 | 1470 |
| AGB28243.1 | 5.70e-291 | 24 | 938 | 546 | 1369 |
| QUT74167.1 | 6.86e-102 | 34 | 931 | 400 | 1426 |
| QCD38476.1 | 9.53e-73 | 439 | 940 | 964 | 1476 |
| QCP72166.1 | 9.53e-73 | 439 | 940 | 964 | 1476 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SMY6 | 1.23e-07 | 526 | 735 | 319 | 528 | Putative pectinesterase/pectinesterase inhibitor 45 OS=Arabidopsis thaliana OX=3702 GN=PME45 PE=2 SV=1 |
| Q47474 | 4.86e-07 | 579 | 722 | 200 | 358 | Pectinesterase B OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemB PE=1 SV=2 |
| Q7Y201 | 1.11e-06 | 527 | 772 | 325 | 571 | Probable pectinesterase/pectinesterase inhibitor 13 OS=Arabidopsis thaliana OX=3702 GN=PME13 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
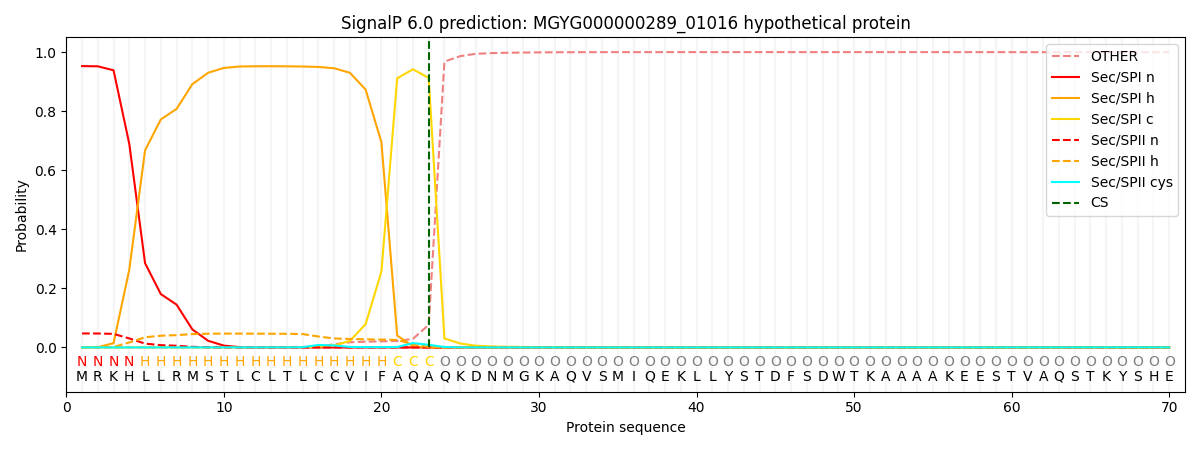
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000739 | 0.948083 | 0.050452 | 0.000262 | 0.000222 | 0.000214 |
