You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000858_01606
You are here: Home > Sequence: MGYG000000858_01606
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Collinsella sp900542155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; Collinsella sp900542155 | |||||||||||
| CAZyme ID | MGYG000000858_01606 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10228; End: 13791 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 366 | 508 | 1.5e-22 | 0.6170212765957447 |
| CBM13 | 213 | 358 | 2.1e-20 | 0.6595744680851063 |
| CBM13 | 808 | 933 | 3.6e-20 | 0.6063829787234043 |
| CBM13 | 900 | 997 | 6.2e-18 | 0.5 |
| CBM13 | 592 | 687 | 1.3e-17 | 0.4946808510638298 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13529 | Peptidase_C39_2 | 2.99e-21 | 1006 | 1159 | 2 | 138 | Peptidase_C39 like family. |
| pfam14200 | RicinB_lectin_2 | 3.95e-21 | 892 | 980 | 2 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.41e-20 | 455 | 545 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 6.48e-19 | 295 | 383 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 7.24e-18 | 852 | 932 | 10 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT49575.1 | 2.24e-192 | 404 | 1187 | 270 | 1061 |
| BAK44091.1 | 2.96e-69 | 248 | 997 | 131 | 885 |
| QRT49576.1 | 2.94e-51 | 397 | 914 | 406 | 928 |
| QOL34941.1 | 9.91e-26 | 102 | 467 | 614 | 978 |
| AMK55176.1 | 1.63e-22 | 100 | 467 | 517 | 884 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5B2H_A | 1.37e-06 | 763 | 895 | 147 | 278 | Crystalstructure of HA33 from Clostridium botulinum serotype C strain Yoichi [Clostridium botulinum],5B2H_B Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi [Clostridium botulinum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34735 | 1.67e-07 | 1001 | 1187 | 59 | 249 | Uncharacterized protein YvpB OS=Bacillus subtilis (strain 168) OX=224308 GN=yvpB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
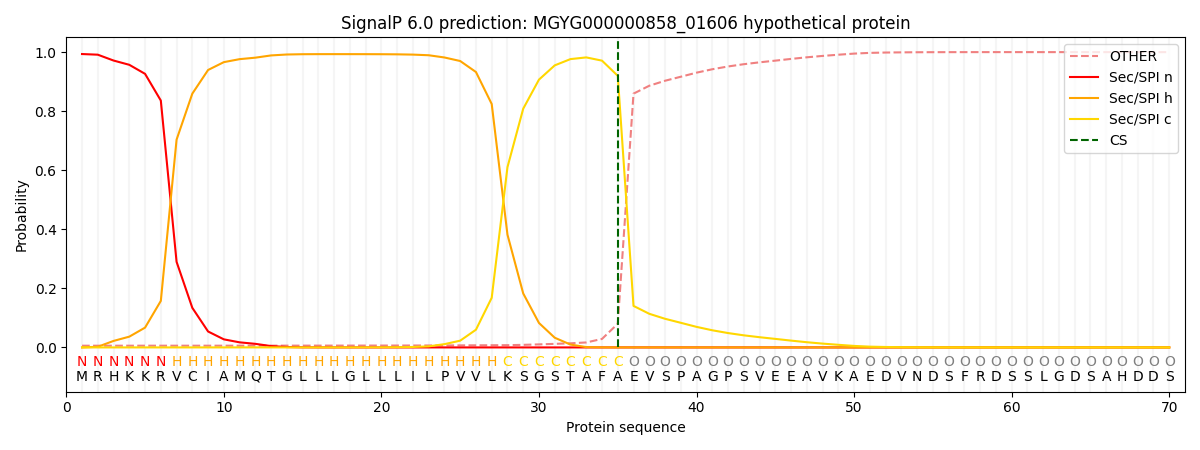
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.007792 | 0.990374 | 0.001074 | 0.000298 | 0.000225 | 0.000214 |