You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000919_03011
You are here: Home > Sequence: MGYG000000919_03011
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Niameybacter sp900551325 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Cellulosilyticaceae; Niameybacter; Niameybacter sp900551325 | |||||||||||
| CAZyme ID | MGYG000000919_03011 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 50101; End: 51732 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 223 | 381 | 1.7e-29 | 0.46864686468646866 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 1.51e-14 | 242 | 409 | 179 | 343 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 4.18e-11 | 229 | 317 | 137 | 230 | Glycosyl hydrolase family 26. |
| sd00006 | TPR | 2.03e-06 | 36 | 92 | 38 | 94 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| sd00006 | TPR | 2.17e-06 | 36 | 102 | 4 | 70 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| COG3063 | PilF | 2.65e-04 | 1 | 102 | 1 | 104 | Tfp pilus assembly protein PilF [Cell motility, Extracellular structures]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADC49280.1 | 2.13e-128 | 34 | 534 | 28 | 514 |
| QPC47538.1 | 3.83e-126 | 1 | 543 | 1 | 524 |
| QKS72465.1 | 3.50e-116 | 1 | 543 | 1 | 522 |
| AEM79915.1 | 4.48e-83 | 132 | 448 | 43 | 341 |
| ADL06956.1 | 1.05e-80 | 135 | 514 | 44 | 411 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2DDX_A | 9.16e-09 | 244 | 356 | 108 | 212 | Crystalstructure of beta-1,3-xylanase from Vibrio sp. AX-4 [Vibrio sp. AX-4],3VPL_A Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase [Vibrio sp.] |
| 3WDQ_A | 1.81e-08 | 231 | 356 | 181 | 299 | Crystalstructure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus [Symbiotic protist of Reticulitermes speratus],3WDR_A Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus complexed with gluco-manno-oligosaccharide [Symbiotic protist of Reticulitermes speratus] |
| 6HF2_A | 1.84e-07 | 217 | 317 | 157 | 263 | Thestructure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus [Bacteroides ovatus ATCC 8483],6HF4_A The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 [Bacteroides ovatus ATCC 8483] |
| 7EET_A | 3.89e-06 | 229 | 318 | 164 | 263 | ChainA, Mannanase KMAN [Klebsiella oxytoca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5MP61 | 9.14e-08 | 244 | 356 | 130 | 234 | Beta-1,3-xylanase XYL4 OS=Vibrio sp. OX=678 GN=xyl4 PE=1 SV=1 |
| P16699 | 2.57e-07 | 228 | 393 | 168 | 321 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| Q9LCB9 | 4.13e-07 | 244 | 382 | 130 | 247 | Beta-1,3-xylanase TXYA OS=Vibrio sp. OX=678 GN=txyA PE=1 SV=1 |
| Q8RS40 | 4.19e-07 | 236 | 382 | 122 | 247 | Beta-1,3-xylanase OS=Alcaligenes sp. OX=512 GN=txyA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
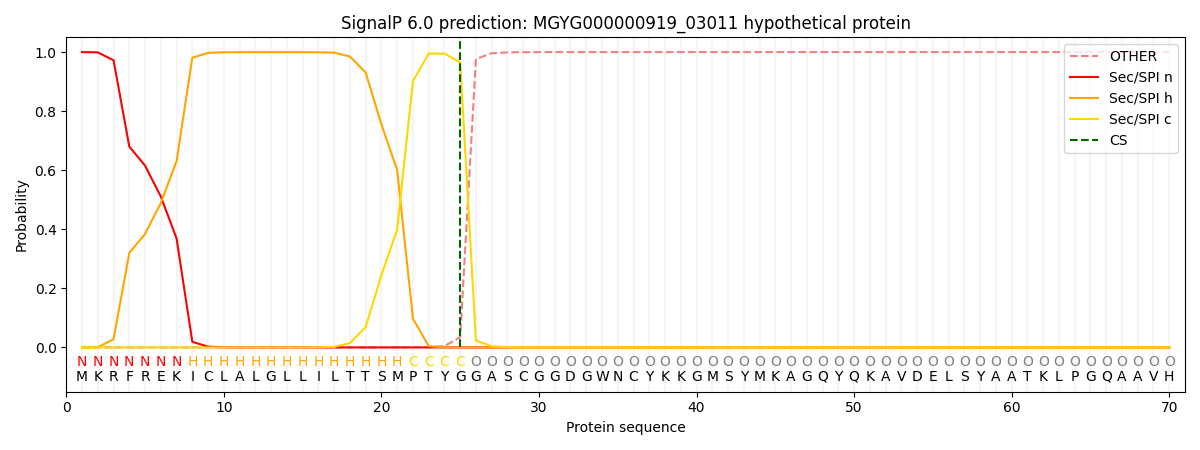
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000482 | 0.998300 | 0.000644 | 0.000187 | 0.000180 | 0.000174 |
