You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001331_01306
Basic Information
help
| Species |
Oxalobacter formigenes
|
| Lineage |
Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Oxalobacter; Oxalobacter formigenes
|
| CAZyme ID |
MGYG000001331_01306
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001331 |
2417984 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 1427904;
End: 1429655
Strand: +
|
No EC number prediction in MGYG000001331_01306.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
27 |
520 |
7.6e-31 |
0.8518518518518519 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
4.02e-16 |
40 |
494 |
23 |
444 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| UAC69887.1
|
1.01e-27 |
37 |
583 |
72 |
625 |
| QDZ27796.1
|
1.23e-06 |
19 |
199 |
66 |
223 |
This protein is predicted as OTHER
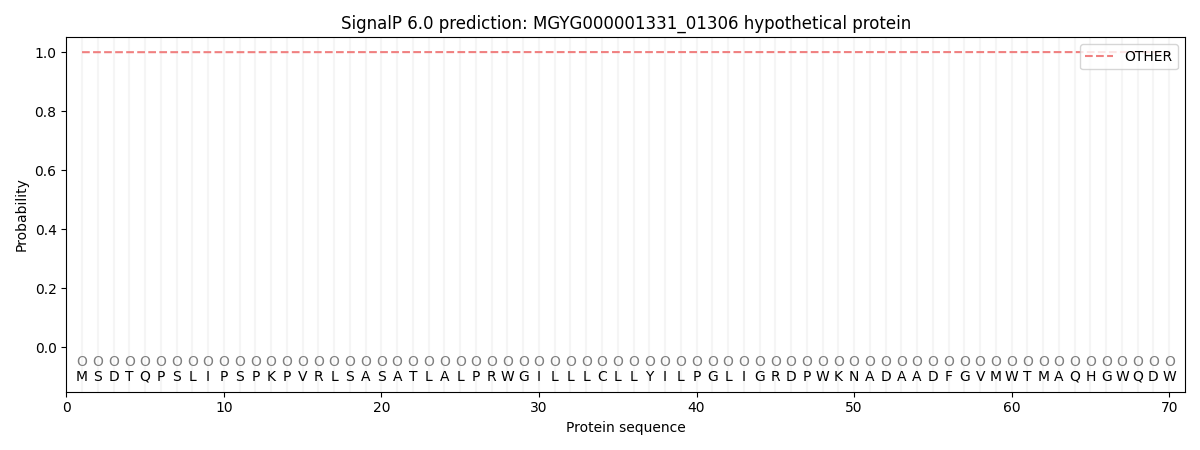
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999907
|
0.000112
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
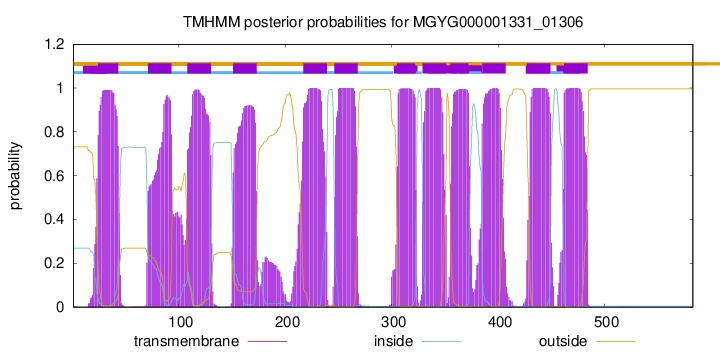
| start |
end |
| 24 |
43 |
| 71 |
93 |
| 108 |
130 |
| 151 |
173 |
| 217 |
239 |
| 246 |
268 |
| 305 |
322 |
| 329 |
351 |
| 355 |
372 |
| 385 |
407 |
| 427 |
449 |
| 462 |
484 |


