You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001385_00771
You are here: Home > Sequence: MGYG000001385_00771
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A indistinctus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A indistinctus | |||||||||||
| CAZyme ID | MGYG000001385_00771 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | Cytoplasmic trehalase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 975320; End: 976732 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH37 | 73 | 453 | 1.4e-71 | 0.7494908350305499 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01204 | Trehalase | 7.19e-61 | 73 | 453 | 128 | 506 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| COG1626 | TreA | 6.67e-52 | 32 | 453 | 120 | 550 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| PRK13270 | treF | 2.01e-33 | 74 | 444 | 173 | 535 | alpha,alpha-trehalase TreF. |
| PLN02567 | PLN02567 | 6.21e-32 | 74 | 398 | 154 | 498 | alpha,alpha-trehalase |
| PRK13272 | treA | 1.43e-31 | 71 | 453 | 159 | 534 | alpha,alpha-trehalase TreA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCG53868.1 | 0.0 | 17 | 470 | 1 | 454 |
| BCG53869.1 | 8.36e-230 | 10 | 456 | 12 | 452 |
| AHM60760.1 | 1.11e-112 | 19 | 444 | 14 | 440 |
| BCI61844.1 | 2.43e-82 | 72 | 453 | 76 | 436 |
| QGA23972.1 | 1.04e-77 | 12 | 453 | 5 | 435 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5N6N_C | 5.47e-32 | 62 | 461 | 297 | 731 | CRYSTALSTRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX [Saccharomyces cerevisiae S288C] |
| 5M4A_A | 5.69e-32 | 62 | 461 | 144 | 578 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae in complex with trehalose [Saccharomyces cerevisiae] |
| 5NIS_A | 7.43e-32 | 62 | 461 | 197 | 631 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae [Saccharomyces cerevisiae S288C] |
| 5JTA_A | 9.81e-32 | 62 | 461 | 292 | 726 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae [Saccharomyces cerevisiae] |
| 2JG0_A | 1.31e-28 | 73 | 455 | 126 | 502 | Family37 trehalase from Escherichia coli in complex with 1- thiatrehazolin [Escherichia coli K-12],2JJB_A Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_B Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_C Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_D Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2WYN_A Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_B Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_C Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_D Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49381 | 6.21e-36 | 73 | 467 | 309 | 734 | Cytosolic neutral trehalase OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=NTH1 PE=1 SV=1 |
| P35172 | 9.05e-36 | 62 | 461 | 321 | 755 | Probable trehalase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=NTH2 PE=1 SV=1 |
| O42893 | 1.96e-32 | 73 | 399 | 287 | 645 | Cytosolic neutral trehalase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ntp1 PE=1 SV=1 |
| J5K1E2 | 3.63e-32 | 73 | 399 | 289 | 657 | Cytosolic neutral trehalase OS=Beauveria bassiana (strain ARSEF 2860) OX=655819 GN=NTH1 PE=1 SV=1 |
| Q2NYS3 | 7.21e-32 | 73 | 457 | 173 | 549 | Periplasmic trehalase OS=Xanthomonas oryzae pv. oryzae (strain MAFF 311018) OX=342109 GN=treA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
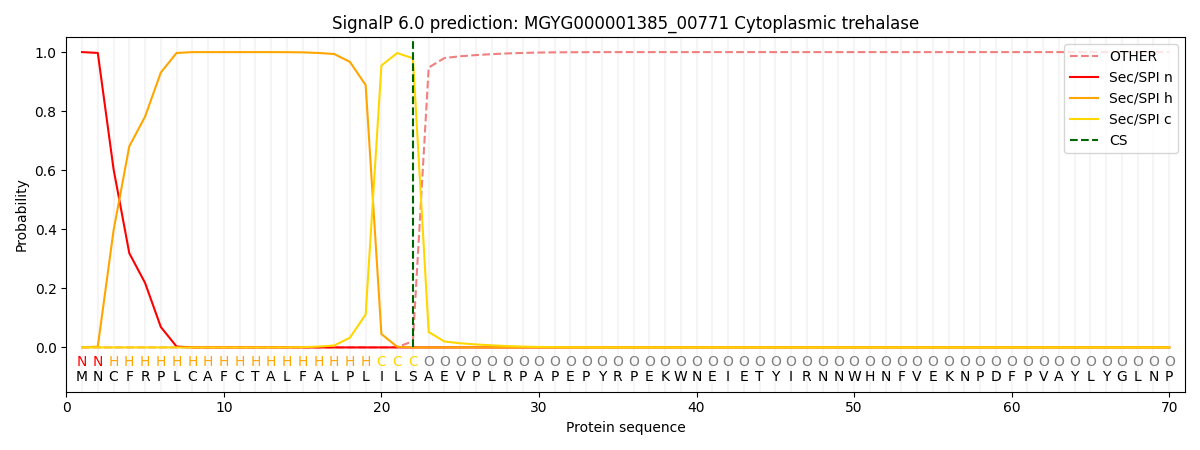
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000224 | 0.999172 | 0.000173 | 0.000154 | 0.000148 | 0.000140 |
