You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001402_01456
You are here: Home > Sequence: MGYG000001402_01456
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
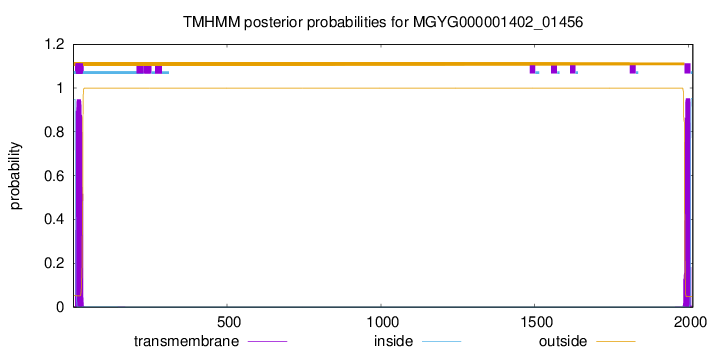
TMHMM annotations
Basic Information help
| Species | Coprobacillus cateniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Coprobacillus; Coprobacillus cateniformis | |||||||||||
| CAZyme ID | MGYG000001402_01456 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1453601; End: 1459645 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 189 | 489 | 6e-96 | 0.9728813559322034 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 1.59e-100 | 189 | 484 | 1 | 288 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 8.59e-20 | 34 | 182 | 1 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 7.10e-07 | 819 | 928 | 10 | 120 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 8.54e-06 | 981 | 1074 | 22 | 128 | Substituted updates: Jan 31, 2002 |
| pfam00754 | F5_F8_type_C | 1.38e-05 | 970 | 1081 | 15 | 115 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM14174.1 | 0.0 | 35 | 1968 | 37 | 1795 |
| BCL58377.1 | 0.0 | 34 | 1825 | 34 | 1791 |
| QTQ18907.1 | 0.0 | 31 | 1972 | 35 | 1836 |
| QMW73896.1 | 0.0 | 30 | 1820 | 48 | 1804 |
| QPS12837.1 | 0.0 | 30 | 1820 | 48 | 1804 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PV4_A | 3.04e-181 | 35 | 649 | 32 | 653 | Structureof CpGH84A [Clostridium perfringens ATCC 13124],6PV4_B Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_C Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_D Structure of CpGH84A [Clostridium perfringens ATCC 13124] |
| 6PWI_A | 1.83e-99 | 34 | 640 | 33 | 624 | Structureof CpGH84D [Clostridium perfringens ATCC 13124],6PWI_B Structure of CpGH84D [Clostridium perfringens ATCC 13124] |
| 6PV5_A | 2.17e-50 | 37 | 634 | 42 | 617 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 5MI4_A | 3.29e-49 | 124 | 741 | 79 | 683 | BtGH84mutant with covalent modification by MA3 [Bacteroides thetaiotaomicron VPI-5482],5MI5_A BtGH84 mutant with covalent modification by MA3 in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482],5MI6_A BtGH84 mutant with covalent modification by MA3 in complex with Thiamet G [Bacteroides thetaiotaomicron VPI-5482],5MI7_A BtGH84 mutant with covalent modification by MA4 in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482] |
| 2V5D_A | 2.12e-48 | 39 | 791 | 18 | 736 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 9.12e-288 | 35 | 1648 | 39 | 1491 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q89ZI2 | 2.08e-47 | 124 | 741 | 89 | 693 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q0TR53 | 6.99e-47 | 39 | 812 | 48 | 782 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 6.99e-47 | 39 | 823 | 48 | 793 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| O60502 | 8.08e-19 | 190 | 449 | 63 | 325 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
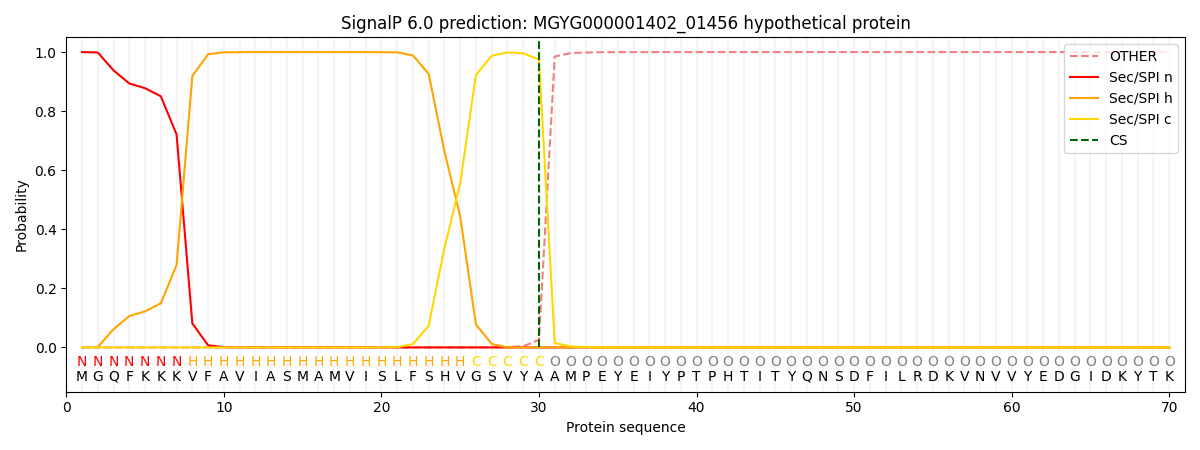
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000261 | 0.999057 | 0.000190 | 0.000168 | 0.000155 | 0.000146 |