You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001404_00166
You are here: Home > Sequence: MGYG000001404_00166
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
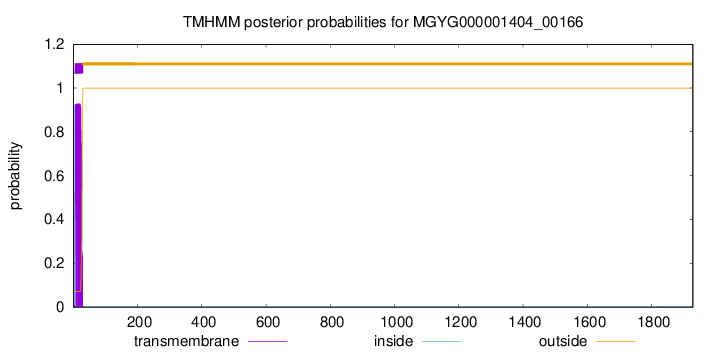
TMHMM annotations
Basic Information help
| Species | Paenibacillus_L senegalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_L; Paenibacillus_L senegalensis | |||||||||||
| CAZyme ID | MGYG000001404_00166 | |||||||||||
| CAZy Family | CBM70 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 167108; End: 172897 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL35 | 813 | 990 | 2.6e-51 | 0.9776536312849162 |
| CBM32 | 1173 | 1289 | 3.7e-24 | 0.9193548387096774 |
| CBM70 | 32 | 191 | 9.4e-24 | 0.9875776397515528 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.14e-17 | 1170 | 1289 | 1 | 126 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| NF033190 | inl_like_NEAT_1 | 2.08e-13 | 1750 | 1921 | 582 | 750 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam02368 | Big_2 | 1.27e-11 | 195 | 270 | 2 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| cd00057 | FA58C | 1.53e-10 | 1164 | 1286 | 7 | 133 | Substituted updates: Jan 31, 2002 |
| COG5492 | YjdB | 2.78e-10 | 175 | 291 | 163 | 279 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACX64738.1 | 0.0 | 28 | 1451 | 32 | 1456 |
| QOT12851.1 | 0.0 | 28 | 1464 | 32 | 1469 |
| AYB44936.1 | 0.0 | 1 | 1464 | 1 | 1467 |
| ANY74958.1 | 0.0 | 2 | 1295 | 6 | 1307 |
| ALS27107.1 | 3.26e-274 | 33 | 1345 | 262 | 1568 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D29_A | 2.27e-30 | 1168 | 1293 | 11 | 132 | CBM32of AlyQ [Persicobacter sp. CCB-QB2],7D2A_A CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid [Persicobacter sp. CCB-QB2] |
| 5XNR_A | 8.56e-28 | 1168 | 1293 | 11 | 132 | TruncatedAlyQ with CBM32 and alginate lyase domains [Persicobacter sp. CCB-QB2] |
| 5ZU6_A | 4.19e-23 | 1156 | 1288 | 17 | 152 | ACBM32 derived from alginate lyase B (AlyB-OU02) [Vibrio] |
| 5ZU5_A | 1.20e-20 | 1156 | 1288 | 17 | 152 | Crystalstructure of a full length alginate lyase with CBM domain [Vibrio splendidus] |
| 2JD9_A | 3.54e-18 | 1168 | 1301 | 10 | 142 | Structureof a pectin binding carbohydrate binding module determined in an orthorhombic crystal form. [Yersinia enterocolitica],2JDA_A Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica],2JDA_B Structure of a pectin binding carbohydrate binding module determined in an monoclinic crystal form. [Yersinia enterocolitica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 1.84e-29 | 1708 | 1926 | 1640 | 1858 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 5.61e-26 | 1757 | 1926 | 914 | 1084 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P19424 | 8.70e-22 | 1752 | 1923 | 43 | 214 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| P49051 | 9.26e-17 | 1748 | 1920 | 32 | 201 | S-layer protein sap OS=Bacillus anthracis OX=1392 GN=sap PE=1 SV=1 |
| C6CRV0 | 9.97e-17 | 1627 | 1919 | 1144 | 1455 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
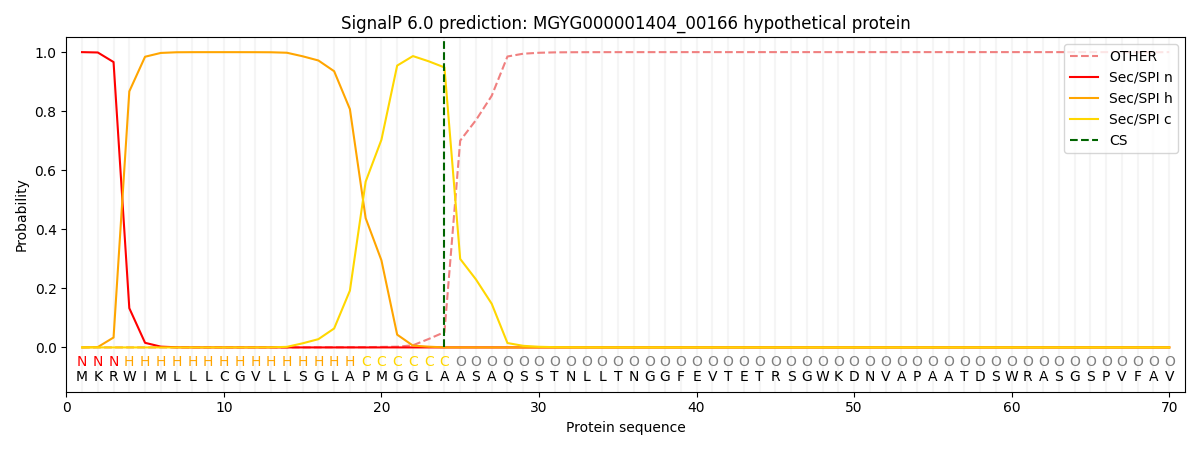
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000775 | 0.998056 | 0.000398 | 0.000333 | 0.000226 | 0.000181 |