You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001657_01234
You are here: Home > Sequence: MGYG000001657_01234
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900760425 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900760425 | |||||||||||
| CAZyme ID | MGYG000001657_01234 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5920; End: 8343 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH105 | 58 | 401 | 2.9e-110 | 0.9668674698795181 |
| PL1 | 481 | 669 | 3.4e-72 | 0.9836065573770492 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 2.96e-95 | 34 | 403 | 1 | 342 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| COG4225 | YesR | 1.50e-93 | 37 | 401 | 9 | 354 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 4.50e-12 | 494 | 675 | 15 | 190 | Amb_all domain. |
| COG3866 | PelB | 8.02e-10 | 441 | 646 | 48 | 246 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam13229 | Beta_helix | 6.02e-05 | 541 | 657 | 24 | 143 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK63671.1 | 1.17e-133 | 3 | 399 | 3 | 395 |
| QUT93209.1 | 4.95e-113 | 21 | 413 | 13 | 406 |
| ALJ61262.1 | 4.95e-113 | 21 | 413 | 13 | 406 |
| QDO69177.1 | 1.38e-112 | 21 | 413 | 13 | 406 |
| BBE19946.1 | 1.53e-111 | 35 | 400 | 33 | 393 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4WU0_A | 2.38e-96 | 61 | 403 | 23 | 361 | StructuralAnalysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 [Clostridium acetobutylicum ATCC 824],4WU0_B Structural Analysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 [Clostridium acetobutylicum ATCC 824] |
| 1NC5_A | 1.30e-56 | 60 | 403 | 38 | 367 | Structureof Protein of Unknown Function of YteR from Bacillus Subtilis [Bacillus subtilis],2D8L_A Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc [Bacillus subtilis] |
| 2GH4_A | 4.98e-56 | 60 | 403 | 28 | 357 | ChainA, Putative glycosyl hydrolase yteR [Bacillus subtilis] |
| 3K11_A | 2.80e-20 | 144 | 408 | 178 | 418 | Crystalstructure of Putative glycosyl hydrolase (NP_813087.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.80 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 3QWT_A | 4.09e-13 | 148 | 331 | 146 | 319 | ChainA, Putative GH105 family protein [Salmonella enterica subsp. enterica serovar Paratyphi A],3QWT_B Chain B, Putative GH105 family protein [Salmonella enterica subsp. enterica serovar Paratyphi A],3QWT_C Chain C, Putative GH105 family protein [Salmonella enterica subsp. enterica serovar Paratyphi A],3QWT_D Chain D, Putative GH105 family protein [Salmonella enterica subsp. enterica serovar Paratyphi A] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34559 | 7.11e-56 | 60 | 403 | 38 | 367 | Unsaturated rhamnogalacturonyl hydrolase YteR OS=Bacillus subtilis (strain 168) OX=224308 GN=yteR PE=1 SV=1 |
| B8NQQ7 | 3.49e-41 | 430 | 784 | 22 | 344 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| B0XMA2 | 3.05e-40 | 424 | 784 | 17 | 345 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
| Q4WL88 | 5.64e-40 | 424 | 784 | 17 | 345 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| A1DPF0 | 5.64e-40 | 424 | 784 | 17 | 345 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
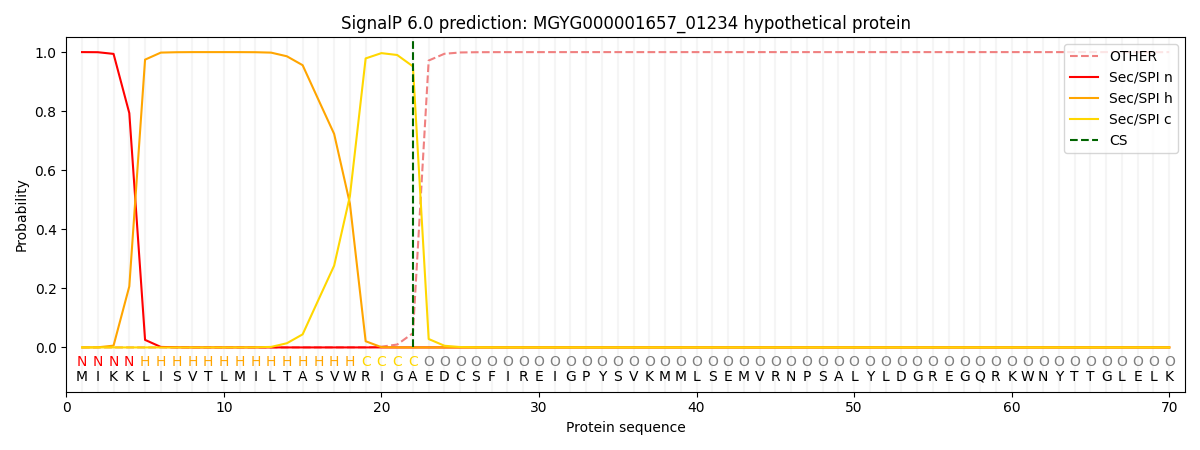
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000475 | 0.998709 | 0.000259 | 0.000185 | 0.000177 | 0.000169 |
