You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002007_01070
You are here: Home > Sequence: MGYG000002007_01070
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes dispar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes dispar | |||||||||||
| CAZyme ID | MGYG000002007_01070 | |||||||||||
| CAZy Family | GH38 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 211373; End: 214822 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH38 | 302 | 574 | 2.9e-18 | 0.9591078066914498 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10791 | GH38N_AMII_like_1 | 2.73e-83 | 302 | 546 | 1 | 254 | N-terminal catalytic domain of mainly uncharacterized eukaryotic proteins similar to alpha-mannosidases; glycoside hydrolase family 38 (GH38). The subfamily of mainly uncharacterized eukaryotic proteins shows sequence homology with class II alpha-mannosidases (AlphaAMIIs). AlphaAMIIs possess a-1,3, a-1,6, and a-1,2 hydrolytic activity, and catalyze the degradation of N-linked oligosaccharides. The N-terminal catalytic domain of alphaMII adopts a structure consisting of parallel 7-stranded beta/alpha barrel. This subfamily belongs to the GH38 family of retaining glycosyl hydrolases, which employ a two-step mechanism involving the formation of a covalent glycosyl enzyme complex; two carboxylic acids positioned within the active site act in concert: one as a catalytic nucleophile and the other as a general acid/base catalyst. |
| pfam01074 | Glyco_hydro_38 | 1.40e-36 | 302 | 577 | 1 | 271 | Glycosyl hydrolases family 38 N-terminal domain. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. |
| pfam14683 | CBM-like | 1.13e-13 | 28 | 191 | 1 | 157 | Polysaccharide lyase family 4, domain III. CBM-like is domain III of rhamnogalacturonan lyase (RG-lyase). The full-length protein specifically recognizes and cleaves alpha-1,4 glycosidic bonds between l-rhamnose and d-galacturonic acids in the backbone of rhamnogalacturonan-I, a major component of the plant cell wall polysaccharide, pectin. This domain possesses a jelly roll beta-sandwich fold structurally homologous to carbohydrate binding modules (CBMs), and it carries two sulfate ions and a hexa-coordinated calcium ion. |
| cd10317 | RGL4_C | 2.45e-11 | 30 | 192 | 1 | 161 | C-terminal domain of rhamnogalacturonan lyase, a family 4 polysaccharide lyase. The rhamnogalacturonan lyase of the polysaccharide lyase family 4 (RGL4) is involved in the degradation of RG (rhamnogalacturonan) type-I, an important pectic plant cell wall polysaccharide, by cleaving the alpha-1,4 glycoside bond between L-rhamnose and D-galacturonic acids in the backbone of RG type-I through a beta-elimination reaction. RGL4 consists of three domains, an N-terminal catalytic domain, a middle domain with a FNIII type fold and a C-terminal domain with a jelly roll fold. Both the middle and the C-terminal domain are putative carbohydrate binding modules. There are two types of RG lyases, which both cleave the alpha-1,4 bonds of the RG-I main chain (RG chain) through the beta-elimination reaction, but belong to two structurally unrelated polysaccharide lyase (PL) families, 4 and 11. |
| pfam07748 | Glyco_hydro_38C | 1.24e-10 | 790 | 968 | 1 | 181 | Glycosyl hydrolases family 38 C-terminal domain. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL07455.1 | 0.0 | 1 | 1149 | 1 | 1149 |
| BBL02110.1 | 0.0 | 1 | 1147 | 1 | 1146 |
| BBL10048.1 | 0.0 | 1 | 1147 | 1 | 1146 |
| BBL12841.1 | 0.0 | 1 | 1147 | 1 | 1146 |
| ALJ40506.1 | 0.0 | 29 | 1149 | 27 | 1140 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
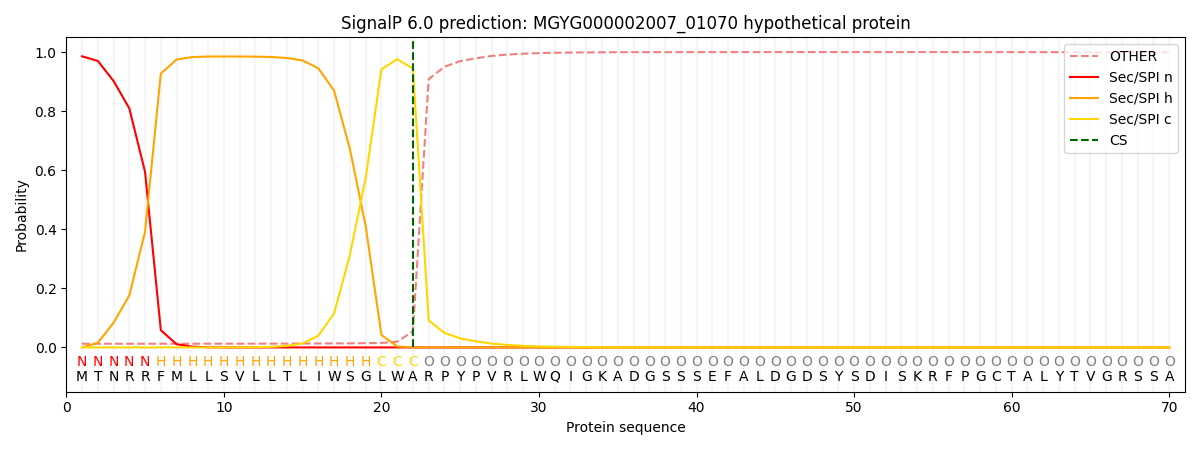
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.016376 | 0.980089 | 0.001245 | 0.001636 | 0.000354 | 0.000270 |
