You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002281_01327
You are here: Home > Sequence: MGYG000002281_01327
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides faecis | |||||||||||
| CAZyme ID | MGYG000002281_01327 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | Arylsulfatase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 218997; End: 220538 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16144 | ARS_like | 2.84e-160 | 33 | 489 | 1 | 420 | uncharacterized arylsulfatase subfamily. Sulfatases catalyze the hydrolysis of sulfate esters from wide range of substrates, including steroids, carbohydrates and proteins. Sulfate esters may be formed from various alcohols and amines. The biological roles of sulfatase includes the cycling of sulfur in the environment, in the degradation of sulfated glycosaminoglycans and glycolipids in the lysosome, and in remodeling sulfated glycosaminoglycans in the extracellular space. The sulfatases are essential for human metabolism. At least eight human monogenic diseases are caused by the deficiency of individual sulfatases. |
| cd16026 | GALNS_like | 1.31e-95 | 32 | 473 | 1 | 399 | galactosamine-6-sulfatase; also known as N-acetylgalactosamine-6-sulfatase (GALNS). Lysosomal galactosamine-6-sulfatase removes sulfate groups from a terminal N-acetylgalactosamine-6-sulfate (or galactose-6-sulfate) in mucopolysaccharides such as keratan sulfate and chondroitin-6-sulfate. Defects in GALNS lead to accumulation of substrates, resulting in the development of the lysosomal storage disease mucopolysaccharidosis IV A. |
| cd16146 | ARS_like | 1.19e-88 | 33 | 489 | 1 | 403 | uncharacterized arylsulfatase. Sulfatases catalyze the hydrolysis of sulfate esters from wide range of substrates, including steroids, carbohydrates and proteins. Sulfate esters may be formed from various alcohols and amines. The biological roles of sulfatase includes the cycling of sulfur in the environment, in the degradation of sulfated glycosaminoglycans and glycolipids in the lysosome, and in remodeling sulfated glycosaminoglycans in the extracellular space. The sulfatases are essential for human metabolism. At least eight human monogenic diseases are caused by the deficiency of individual sulfatases. |
| cd16029 | 4-S | 1.72e-79 | 33 | 473 | 1 | 393 | N-acetylgalactosamine 4-sulfatase, also called arylsulftase B. Sulfatases catalyze the hydrolysis of sulfuric acid esters from a wide variety of substrates. N-acetylgalactosamine 4-sulfatase catalyzes the removal of the sulfate ester group from position 4 of an N-acetylgalactosamine sugar at the non-reducing terminus of the polysaccharide in the degradative pathways of the glycosaminoglycans dermatan sulfate and chondroitin-4-sulfate. N-acetylgalactosamine 4-sulfatase is a lysosomal enzyme. |
| cd16143 | ARS_like | 8.83e-79 | 33 | 473 | 1 | 395 | uncharacterized arylsulfatase subfamily. Sulfatases catalyze the hydrolysis of sulfate esters from wide range of substrates, including steroids, carbohydrates and proteins. Sulfate esters may be formed from various alcohols and amines. The biological roles of sulfatase includes the cycling of sulfur in the environment, in the degradation of sulfated glycosaminoglycans and glycolipids in the lysosome, and in remodeling sulfated glycosaminoglycans in the extracellular space. The sulfatases are essential for human metabolism. At least eight human monogenic diseases are caused by the deficiency of individual sulfatases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT89376.1 | 5.00e-106 | 28 | 510 | 19 | 496 |
| ALJ59588.1 | 3.82e-105 | 28 | 510 | 19 | 496 |
| EAR02039.1 | 4.38e-100 | 28 | 510 | 24 | 507 |
| AKJ65363.1 | 2.61e-57 | 31 | 499 | 25 | 452 |
| QDT74124.1 | 1.04e-41 | 17 | 491 | 21 | 463 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6USS_A | 5.81e-106 | 27 | 499 | 28 | 502 | ChainA, Sulfatase [Bacteroides fragilis CAG:558],6USS_B Chain B, Sulfatase [Bacteroides fragilis CAG:558] |
| 6UST_A | 8.52e-46 | 32 | 478 | 4 | 440 | ChainA, N-acetylgalactosamine 6-sulfate sulfatase [Hungatella hathewayi],6UST_B Chain B, N-acetylgalactosamine 6-sulfate sulfatase [Hungatella hathewayi],6UST_C Chain C, N-acetylgalactosamine 6-sulfate sulfatase [Hungatella hathewayi],6UST_D Chain D, N-acetylgalactosamine 6-sulfate sulfatase [Hungatella hathewayi] |
| 6PSM_A | 1.60e-45 | 31 | 487 | 23 | 433 | Crystalstructure of PsS1_19B C77S in complex with kappa-neocarrabiose [Pseudoalteromonas fuliginea],6PSM_B Crystal structure of PsS1_19B C77S in complex with kappa-neocarrabiose [Pseudoalteromonas fuliginea],6PSM_C Crystal structure of PsS1_19B C77S in complex with kappa-neocarrabiose [Pseudoalteromonas fuliginea],6PSM_D Crystal structure of PsS1_19B C77S in complex with kappa-neocarrabiose [Pseudoalteromonas fuliginea],6PSM_E Crystal structure of PsS1_19B C77S in complex with kappa-neocarrabiose [Pseudoalteromonas fuliginea],6PSM_F Crystal structure of PsS1_19B C77S in complex with kappa-neocarrabiose [Pseudoalteromonas fuliginea],6PSO_A Crystal structure of PsS1_19B C77S in complex with iota-neocarratetraose [Pseudoalteromonas fuliginea],6PSO_B Crystal structure of PsS1_19B C77S in complex with iota-neocarratetraose [Pseudoalteromonas fuliginea] |
| 6PRM_A | 8.13e-45 | 31 | 487 | 23 | 433 | Crystalstructure of apo PsS1_19B [Pseudoalteromonas fuliginea],6PRM_B Crystal structure of apo PsS1_19B [Pseudoalteromonas fuliginea],6PRM_C Crystal structure of apo PsS1_19B [Pseudoalteromonas fuliginea],6PRM_D Crystal structure of apo PsS1_19B [Pseudoalteromonas fuliginea] |
| 6B0K_A | 1.81e-40 | 31 | 487 | 1 | 415 | Crystalstructure of Ps i-CgsB C78S in complex with k-carrapentaose [Pseudoalteromonas],6B0K_B Crystal structure of Ps i-CgsB C78S in complex with k-carrapentaose [Pseudoalteromonas],6B0K_C Crystal structure of Ps i-CgsB C78S in complex with k-carrapentaose [Pseudoalteromonas] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q32KH5 | 1.35e-41 | 17 | 442 | 15 | 399 | N-acetylgalactosamine-6-sulfatase OS=Canis lupus familiaris OX=9615 GN=GALNS PE=2 SV=1 |
| Q571E4 | 2.48e-41 | 17 | 442 | 13 | 397 | N-acetylgalactosamine-6-sulfatase OS=Mus musculus OX=10090 GN=Galns PE=1 SV=2 |
| Q32KJ6 | 1.78e-40 | 17 | 404 | 17 | 370 | N-acetylgalactosamine-6-sulfatase OS=Rattus norvegicus OX=10116 GN=Galns PE=1 SV=1 |
| Q8WNQ7 | 1.07e-38 | 17 | 442 | 15 | 399 | N-acetylgalactosamine-6-sulfatase OS=Sus scrofa OX=9823 GN=GALNS PE=2 SV=1 |
| P34059 | 2.01e-38 | 29 | 404 | 27 | 368 | N-acetylgalactosamine-6-sulfatase OS=Homo sapiens OX=9606 GN=GALNS PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
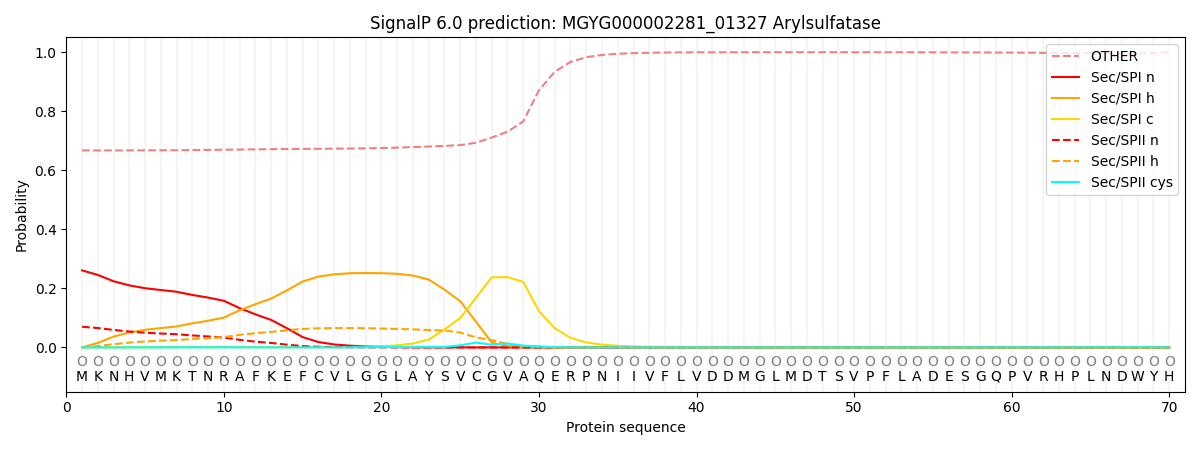
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.680437 | 0.242086 | 0.072754 | 0.002282 | 0.001067 | 0.001377 |
