You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002291_02600
You are here: Home > Sequence: MGYG000002291_02600
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
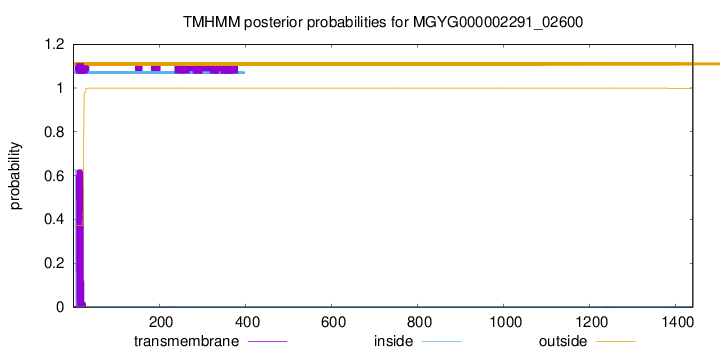
TMHMM annotations
Basic Information help
| Species | OM05-12 sp003438995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; OM05-12; OM05-12 sp003438995 | |||||||||||
| CAZyme ID | MGYG000002291_02600 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20479; End: 24801 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 828 | 1033 | 7.4e-35 | 0.9722222222222222 |
| CBM32 | 258 | 372 | 2.5e-25 | 0.9032258064516129 |
| CBM32 | 500 | 614 | 1.1e-18 | 0.8951612903225806 |
| CBM32 | 33 | 151 | 9.5e-18 | 0.9112903225806451 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 1.92e-43 | 780 | 1351 | 35 | 650 | beta-glucosidase BglX. |
| COG1472 | BglX | 2.82e-43 | 790 | 1141 | 1 | 368 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 5.08e-28 | 817 | 1069 | 47 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 5.09e-28 | 1110 | 1312 | 1 | 213 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| pfam00754 | F5_F8_type_C | 1.36e-19 | 255 | 369 | 2 | 123 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCB77526.1 | 7.74e-300 | 217 | 1349 | 13 | 1040 |
| BCJ74909.1 | 6.56e-299 | 252 | 1349 | 48 | 1042 |
| SBT46049.1 | 6.95e-294 | 212 | 1349 | 11 | 1027 |
| BCB91177.1 | 4.24e-293 | 212 | 1349 | 11 | 1040 |
| QLQ36641.1 | 2.60e-291 | 257 | 1349 | 55 | 1029 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M6G_A | 9.66e-45 | 772 | 1301 | 40 | 602 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 4ZOA_A | 2.59e-36 | 782 | 1293 | 5 | 561 | CrystalStructure of beta-glucosidase from Listeria innocua in complex with isofagomine [Listeria innocua Clip11262],4ZOA_B Crystal Structure of beta-glucosidase from Listeria innocua in complex with isofagomine [Listeria innocua Clip11262],4ZOB_A Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone [Listeria innocua Clip11262],4ZOB_B Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone [Listeria innocua Clip11262],4ZOD_A Crystal Structure of beta-glucosidase from Listeria innocua in complex with glucose [Listeria innocua Clip11262],4ZOD_B Crystal Structure of beta-glucosidase from Listeria innocua in complex with glucose [Listeria innocua Clip11262],4ZOE_A Crystal Structure of beta-glucosidase from Listeria innocua [Listeria innocua Clip11262],4ZOE_B Crystal Structure of beta-glucosidase from Listeria innocua [Listeria innocua Clip11262] |
| 5XXL_A | 9.62e-36 | 779 | 1351 | 10 | 637 | Crystalstructure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXL_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXM_A Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482],5XXM_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482] |
| 6R5I_A | 1.44e-35 | 810 | 1293 | 47 | 574 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 4ZO6_A | 1.88e-35 | 782 | 1293 | 5 | 561 | CrystalStructure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with cellobiose [Listeria innocua Clip11262],4ZO6_B Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with cellobiose [Listeria innocua Clip11262],4ZO7_A Crystal structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with gentiobiose [Listeria innocua Clip11262],4ZO7_B Crystal structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with gentiobiose [Listeria innocua Clip11262],4ZO8_A Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorose [Listeria innocua Clip11262],4ZO8_B Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorose [Listeria innocua Clip11262],4ZO9_A Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with laminaribiose [Listeria innocua Clip11262],4ZO9_B Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with laminaribiose [Listeria innocua Clip11262],4ZOC_A Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorotriose [Listeria innocua Clip11262],4ZOC_B Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorotriose [Listeria innocua Clip11262] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5BCC6 | 9.45e-45 | 741 | 1347 | 5 | 614 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| Q46684 | 6.70e-44 | 761 | 1346 | 47 | 652 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| Q2UFP8 | 3.36e-41 | 765 | 1310 | 39 | 612 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| B8NGU6 | 5.74e-41 | 772 | 1310 | 42 | 608 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q56078 | 1.99e-38 | 780 | 1293 | 35 | 606 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
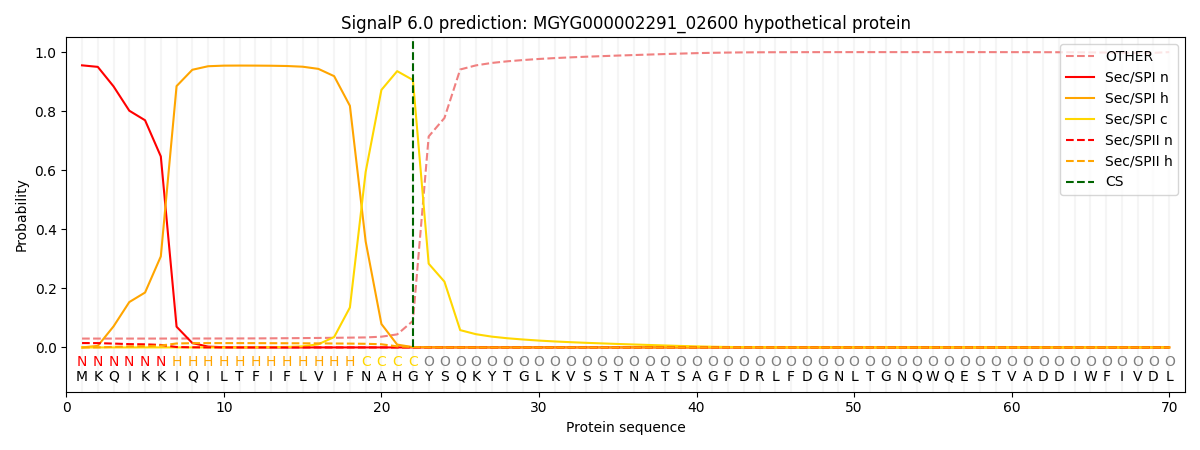
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.037776 | 0.944838 | 0.016130 | 0.000463 | 0.000359 | 0.000399 |