You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002306_04364
You are here: Home > Sequence: MGYG000002306_04364
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mycobacterium avium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium avium | |||||||||||
| CAZyme ID | MGYG000002306_04364 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | Diacylglycerol acyltransferase/mycolyltransferase Ag85A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3425; End: 4468 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 73 | 321 | 4.1e-45 | 0.8810572687224669 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0627 | FrmB | 1.56e-93 | 5 | 325 | 1 | 312 | S-formylglutathione hydrolase FrmB [Defense mechanisms]. |
| pfam00756 | Esterase | 7.80e-79 | 58 | 321 | 1 | 246 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| COG0412 | DLH | 0.007 | 151 | 230 | 102 | 178 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZB12632.1 | 7.74e-258 | 1 | 347 | 1 | 347 |
| QPM69797.1 | 7.74e-258 | 1 | 347 | 1 | 347 |
| AGL38514.1 | 7.74e-258 | 1 | 347 | 1 | 347 |
| QQK48673.1 | 7.74e-258 | 1 | 347 | 1 | 347 |
| ASF94534.1 | 7.74e-258 | 1 | 347 | 1 | 347 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1SFR_A | 1.67e-188 | 43 | 334 | 1 | 292 | ChainA, Antigen 85-A [Mycobacterium tuberculosis],1SFR_B Chain B, Antigen 85-A [Mycobacterium tuberculosis],1SFR_C Chain C, Antigen 85-A [Mycobacterium tuberculosis] |
| 1F0N_A | 1.78e-183 | 44 | 327 | 1 | 284 | ChainA, ANTIGEN 85B [Mycobacterium tuberculosis],1F0P_A Chain A, ANTIGEN 85-B [Mycobacterium tuberculosis] |
| 1VA5_A | 7.90e-147 | 44 | 333 | 2 | 290 | ChainA, Antigen 85-C [Mycobacterium tuberculosis],1VA5_B Chain B, Antigen 85-C [Mycobacterium tuberculosis] |
| 4MQM_A | 2.34e-146 | 43 | 329 | 2 | 287 | ChainA, Diacylglycerol acyltransferase/mycolyltransferase Ag85C [Mycobacterium tuberculosis] |
| 4QEK_A | 6.01e-146 | 43 | 329 | 1 | 286 | Crystalstructure of Antigen 85C-S124A mutant [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O52956 | 8.96e-258 | 1 | 347 | 1 | 347 | Diacylglycerol acyltransferase/mycolyltransferase Ag85A OS=Mycobacterium avium OX=1764 GN=fbpA PE=3 SV=1 |
| O06052 | 2.75e-225 | 1 | 339 | 1 | 338 | Diacylglycerol acyltransferase/mycolyltransferase Ag85A OS=Mycobacterium gordonae OX=1778 GN=fbpA PE=3 SV=1 |
| P58248 | 1.16e-221 | 1 | 332 | 1 | 332 | Diacylglycerol acyltransferase/mycolyltransferase Ag85A OS=Mycobacterium ulcerans OX=1809 GN=fbpA PE=3 SV=1 |
| P9WQP3 | 3.21e-210 | 1 | 334 | 1 | 334 | Diacylglycerol acyltransferase/mycolyltransferase Ag85A OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=fbpA PE=1 SV=1 |
| P0C2T1 | 3.21e-210 | 1 | 334 | 1 | 334 | Diacylglycerol acyltransferase/mycolyltransferase Ag85A OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=fbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
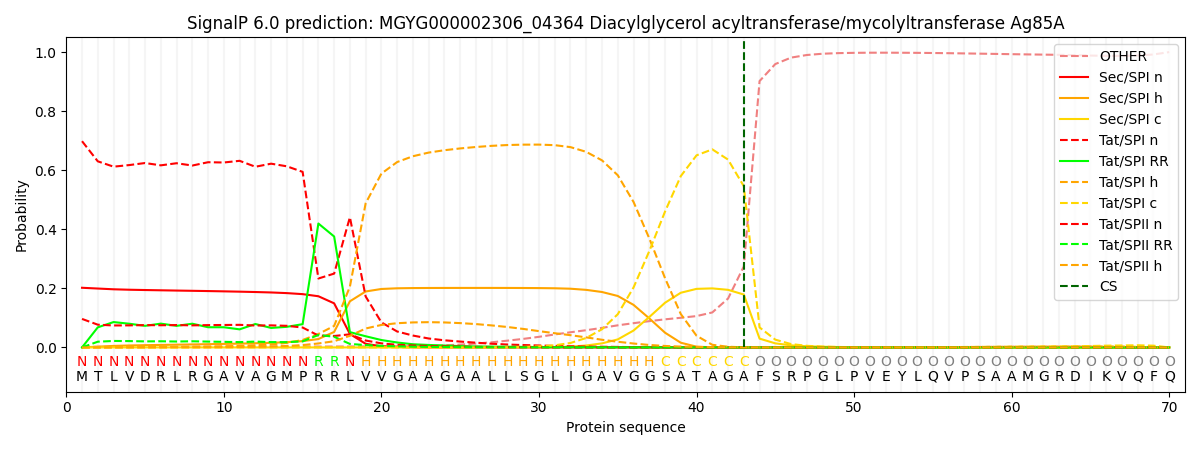
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004229 | 0.189226 | 0.001813 | 0.705412 | 0.098675 | 0.000631 |

