You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002321_00657
You are here: Home > Sequence: MGYG000002321_00657
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
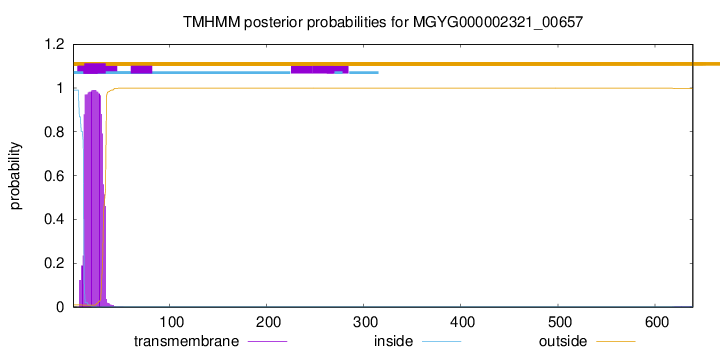
TMHMM annotations
Basic Information help
| Species | Ruminococcus_D bicirculans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_D; Ruminococcus_D bicirculans | |||||||||||
| CAZyme ID | MGYG000002321_00657 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 713242; End: 715161 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 49 | 295 | 2.3e-97 | 0.9881422924901185 |
| CBM65 | 521 | 635 | 1.1e-27 | 0.9649122807017544 |
| CBM13 | 356 | 453 | 1.4e-18 | 0.4946808510638298 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.48e-36 | 48 | 292 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| pfam18259 | CBM65_1 | 6.13e-25 | 522 | 635 | 1 | 113 | Carbohydrate binding module 65 domain 1. This domain is found in the non-catalytic carbohydrate binding module 65B (CMB65B) present in Eubacterium cellulosolvens. CBMs are present in plant cell wall degrading enzymes and are responsible for targeting, which enhances catalysis. CBM65s display higher affinity for oligosaccharides, such as cellohexaose, and particularly polysaccharides than cellotetraose, which fully occupies the core component of the substrate binding cleft. The concave surface presented by beta-sheet 2 comprises the beta-glucan binding site in CBM65s. C6 of all the backbone glucose moieties makes extensive hydrophobic interactions with the surface tryptophans of CBM65s. Three out of the four surface Trp are highly conserved. The conserved metal ion site typical of CBMs is absent in this CBM65 family. |
| pfam14200 | RicinB_lectin_2 | 2.34e-19 | 359 | 438 | 12 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 3.28e-18 | 395 | 486 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 7.10e-08 | 361 | 496 | 2 | 126 | Ricin-type beta-trefoil lectin domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO04360.1 | 0.0 | 1 | 639 | 1 | 639 |
| ADU21877.1 | 2.78e-207 | 4 | 639 | 3 | 485 |
| ADU23074.1 | 3.50e-151 | 20 | 335 | 15 | 331 |
| BAA25878.1 | 3.97e-137 | 31 | 335 | 29 | 333 |
| BAD99527.1 | 6.84e-137 | 31 | 338 | 27 | 334 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1WKY_A | 3.39e-137 | 34 | 338 | 4 | 308 | Crystalstructure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module [Bacillus sp. JAMB-602] |
| 3JUG_A | 1.33e-123 | 27 | 336 | 12 | 321 | Crystalstructure of endo-beta-1,4-mannanase from the alkaliphilic Bacillus sp. N16-5 [Bacillus sp. N16-5] |
| 2WHJ_A | 6.67e-122 | 38 | 333 | 1 | 296 | Understandinghow diverse mannanases recognise heterogeneous substrates [Salipaludibacillus agaradhaerens] |
| 2WHL_A | 1.86e-117 | 39 | 333 | 1 | 293 | Understandinghow diverse mannanases recognise heterogeneous substrates [Salipaludibacillus agaradhaerens] |
| 1BQC_A | 1.69e-79 | 37 | 333 | 1 | 301 | Beta-MannanaseFrom Thermomonospora Fusca [Thermobifida fusca],2MAN_A Mannotriose Complex Of Thermomonospora Fusca Beta-Mannanase [Thermobifida fusca],3MAN_A Mannohexaose Complex Of Thermomonospora Fusca Beta-mannanase [Thermobifida fusca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G1K3N4 | 3.65e-121 | 38 | 333 | 1 | 296 | Mannan endo-1,4-beta-mannosidase OS=Salipaludibacillus agaradhaerens OX=76935 PE=1 SV=1 |
| P51529 | 2.43e-89 | 10 | 339 | 6 | 341 | Mannan endo-1,4-beta-mannosidase OS=Streptomyces lividans OX=1916 GN=manA PE=1 SV=2 |
| B3PF24 | 1.56e-82 | 23 | 340 | 33 | 352 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=man5A PE=1 SV=1 |
| P22533 | 3.48e-60 | 59 | 313 | 55 | 313 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
| P07983 | 7.10e-10 | 19 | 287 | 13 | 292 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
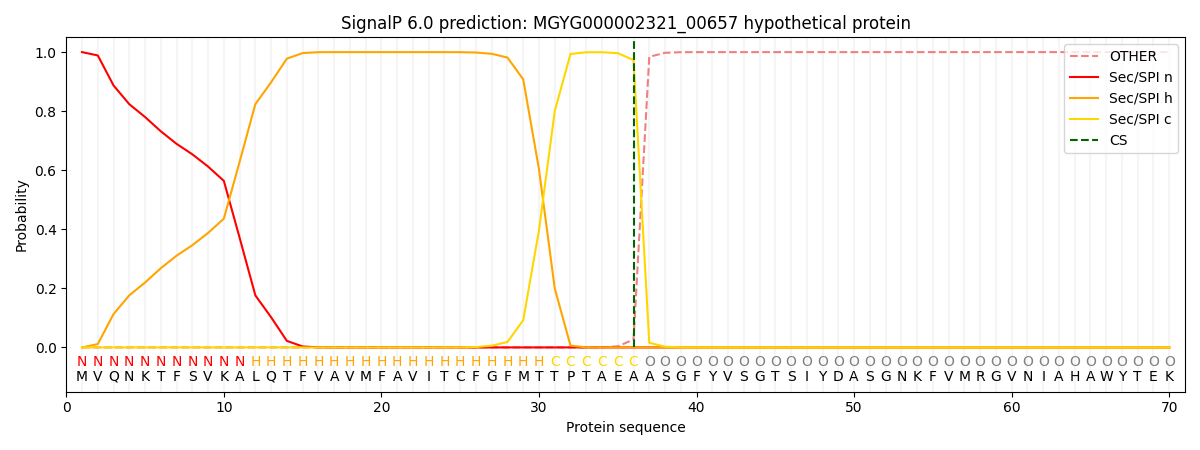
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000284 | 0.998958 | 0.000204 | 0.000187 | 0.000167 | 0.000153 |