You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002359_02155
You are here: Home > Sequence: MGYG000002359_02155
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mycobacterium tuberculosis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium tuberculosis | |||||||||||
| CAZyme ID | MGYG000002359_02155 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Bifunctional apolipoprotein N-acyltransferase/polyprenol monophosphomannose synthase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12316; End: 15105 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 669 | 834 | 6.3e-30 | 0.9764705882352941 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0815 | Lnt | 3.84e-122 | 64 | 590 | 1 | 518 | Apolipoprotein N-acyltransferase [Cell wall/membrane/envelope biogenesis]. |
| TIGR00546 | lnt | 3.46e-111 | 135 | 532 | 5 | 389 | apolipoprotein N-acyltransferase. This enzyme transfers the acyl group to lipoproteins in the lgt/lsp/lnt system which is found broadly in bacteria but not in archaea. This model represents one component of the "lipoprotein lgt/lsp/lnt system" genome property. [Protein fate, Protein modification and repair] |
| cd06442 | DPM1_like | 1.54e-96 | 669 | 894 | 1 | 224 | DPM1_like represents putative enzymes similar to eukaryotic DPM1. Proteins similar to eukaryotic DPM1, including enzymes from bacteria and archaea; DPM1 is the catalytic subunit of eukaryotic dolichol-phosphate mannose (DPM) synthase. DPM synthase is required for synthesis of the glycosylphosphatidylinositol (GPI) anchor, N-glycan precursor, protein O-mannose, and C-mannose. In higher eukaryotes,the enzyme has three subunits, DPM1, DPM2 and DPM3. DPM is synthesized from dolichol phosphate and GDP-Man on the cytosolic surface of the ER membrane by DPM synthase and then is flipped onto the luminal side and used as a donor substrate. In lower eukaryotes, such as Saccharomyces cerevisiae and Trypanosoma brucei, DPM synthase consists of a single component (Dpm1p and TbDpm1, respectively) that possesses one predicted transmembrane region near the C terminus for anchoring to the ER membrane. In contrast, the Dpm1 homologues of higher eukaryotes, namely fission yeast, fungi, and animals, have no transmembrane region, suggesting the existence of adapter molecules for membrane anchoring. This family also includes bacteria and archaea DPM1_like enzymes. However, the enzyme structure and mechanism of function are not well understood. This protein family belongs to Glycosyltransferase 2 superfamily. |
| cd07571 | ALP_N-acyl_transferase | 2.16e-85 | 296 | 569 | 1 | 266 | Apolipoprotein N-acyl transferase (class 9 nitrilases). ALP N-acyl transferase (Lnt), is an essential membrane-bound enzyme in gram-negative bacteria, which catalyzes the N-acylation of apolipoproteins, the final step in lipoprotein maturation. This is a reverse amidase (i.e. condensation) reaction. This subgroup belongs to a larger nitrilase superfamily comprised of nitrile- or amide-hydrolyzing enzymes and amide-condensing enzymes, which depend on a Glu-Lys-Cys catalytic triad. This superfamily has been classified in the literature based on global and structure based sequence analysis into thirteen different enzyme classes (referred to as 1-13), this subgroup corresponds to class 9. |
| PRK00302 | lnt | 1.52e-55 | 74 | 569 | 3 | 486 | apolipoprotein N-acyltransferase; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAL66057.1 | 0.0 | 1 | 929 | 1 | 929 |
| AMC46469.1 | 0.0 | 1 | 929 | 1 | 929 |
| AOE36439.1 | 0.0 | 1 | 929 | 1 | 929 |
| ACT24998.1 | 0.0 | 1 | 929 | 1 | 929 |
| AHJ55258.1 | 0.0 | 1 | 929 | 1 | 929 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MLZ_A | 9.46e-37 | 664 | 902 | 22 | 255 | Dolichylphosphate mannose synthase in complex with GDP and Mg2+ [Pyrococcus furiosus DSM 3638],5MM0_A Dolichyl phosphate mannose synthase in complex with GDP-mannose and Mn2+ (donor complex) [Pyrococcus furiosus DSM 3638],5MM1_A Dolichyl phosphate mannose synthase in complex with GDP and dolichyl phosphate mannose [Pyrococcus furiosus DSM 3638] |
| 5XHQ_A | 1.40e-29 | 79 | 583 | 13 | 508 | ApolipoproteinN-acyl Transferase [Escherichia coli K-12],5XHQ_B Apolipoprotein N-acyl Transferase [Escherichia coli K-12] |
| 6Q3A_A | 1.05e-28 | 79 | 584 | 15 | 511 | Apoform of Apolipoprotein N-acyltransferase (Lnt) [Escherichia coli K-12] |
| 5VRG_A | 1.14e-28 | 79 | 584 | 15 | 511 | Structuralinsights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase [Escherichia coli K-12] |
| 6NWR_A | 1.20e-28 | 79 | 584 | 13 | 509 | Thioesteracyl-intermediate of Apolipoprotein N-acyltransferase (Lnt) [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A0H3M5A8 | 0.0 | 56 | 929 | 1 | 874 | Bifunctional apolipoprotein N-acyltransferase/polyprenol monophosphomannose synthase OS=Mycobacterium bovis (strain BCG / Pasteur 1173P2) OX=410289 GN=lnt PE=1 SV=1 |
| O53493 | 0.0 | 56 | 929 | 1 | 874 | Bifunctional apolipoprotein N-acyltransferase/polyprenol monophosphomannose synthase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=ppm1 PE=1 SV=1 |
| A0QZ13 | 6.33e-214 | 57 | 588 | 47 | 579 | Apolipoprotein N-acyltransferase OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=lnt PE=1 SV=2 |
| A0QZ12 | 1.79e-135 | 662 | 903 | 19 | 260 | Polyprenol monophosphomannose synthase OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=ppm1 PE=1 SV=1 |
| C5CBV8 | 2.27e-84 | 666 | 899 | 2 | 233 | Undecaprenyl-phosphate mannosyltransferase OS=Micrococcus luteus (strain ATCC 4698 / DSM 20030 / JCM 1464 / NBRC 3333 / NCIMB 9278 / NCTC 2665 / VKM Ac-2230) OX=465515 GN=Mlut_12000 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000062 | 0.000004 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
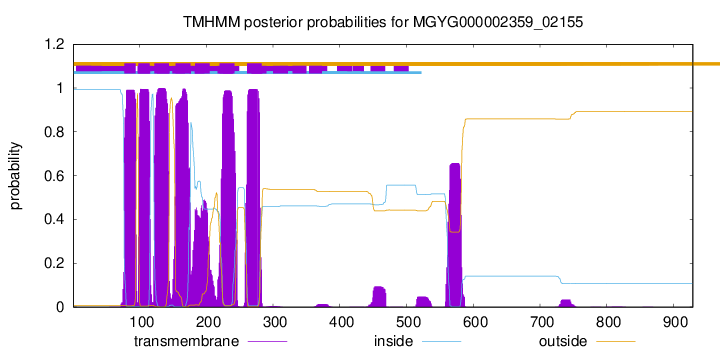
| start | end |
|---|---|
| 77 | 94 |
| 98 | 115 |
| 122 | 144 |
| 154 | 176 |
| 183 | 205 |
| 220 | 242 |
| 261 | 283 |
