You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002379_01106
You are here: Home > Sequence: MGYG000002379_01106
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
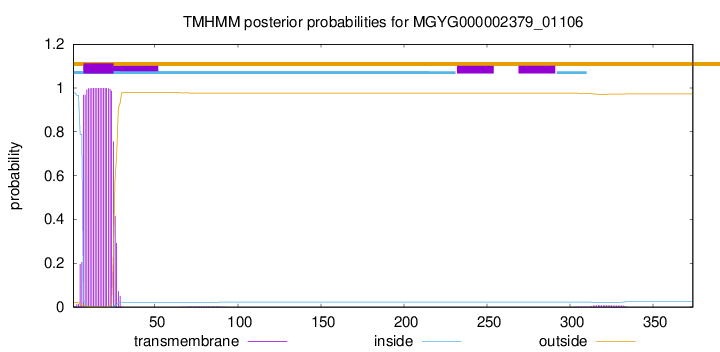
TMHMM annotations
Basic Information help
| Species | Lactobacillus acidophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactobacillus; Lactobacillus acidophilus | |||||||||||
| CAZyme ID | MGYG000002379_01106 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16590; End: 17714 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3405 | BcsZ | 1.64e-22 | 12 | 369 | 2 | 342 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| pfam01270 | Glyco_hydro_8 | 3.76e-22 | 38 | 325 | 8 | 277 | Glycosyl hydrolases family 8. |
| PRK11097 | PRK11097 | 6.51e-14 | 69 | 364 | 54 | 345 | cellulase. |
| pfam05147 | LANC_like | 4.66e-05 | 60 | 230 | 154 | 303 | Lanthionine synthetase C-like protein. Lanthionines are thioether bridges that are putatively generated by dehydration of Ser and Thr residues followed by addition of cysteine residues within the peptide. This family contains the lanthionine synthetase C-like proteins 1 and 2 which are related to the bacterial lanthionine synthetase components C (LanC). LANCL1 (P40 seven-transmembrane-domain protein) and LANCL2 (testes-specific adriamycin sensitivity protein) are thought to be peptide-modifying enzyme components in eukaryotic cells. Both proteins are produced in large quantities in the brain and testes and may have role in the immune surveillance of these organs. Lanthionines are found in lantibiotics, which are peptide-derived, post-translationally modified antimicrobials produced by several bacterial strains. This region contains seven internal repeats. |
| cd04793 | LanC | 0.001 | 80 | 174 | 184 | 283 | Cyclases involved in the biosynthesis of lantibiotics. LanC is the cyclase enzyme of the lanthionine synthetase. Lanthinoine is a lantibiotic, a unique class of peptide antibiotics. They are ribosomally synthesized as precursor peptides and then post-translationally modified to contain thioether cross-links called lanthionines (Lans) or methyllanthionines (MeLans) in addition to 2,3-didehydroalanine (Dha) and (Z)-2,3-didehydrobutyrine (Dhb). These unusual amino acids are introduced by the dehydration of serine and threonine residues, followed by thioether formation via addition of cysteine thiols, catalysed by LanB and LanC or LanM. LanC, the cyclase component, is a zinc metalloprotein, whose bound metal has been proposed to activate the thiol substrate for nucleophilic addition. Also contains SpaC (the cyclase involved in the biosynthesis of subtilin), NisC, and homologs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASN46048.1 | 2.56e-272 | 1 | 374 | 1 | 374 |
| AAV42008.1 | 2.56e-272 | 1 | 374 | 1 | 374 |
| ASX14127.1 | 2.56e-272 | 1 | 374 | 1 | 374 |
| AVW87761.1 | 2.56e-272 | 1 | 374 | 1 | 374 |
| AGK93337.1 | 2.56e-272 | 1 | 374 | 1 | 374 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XD0_A | 1.96e-28 | 27 | 325 | 58 | 359 | ApoStructure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4],5XD0_B Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4] |
| 1V5C_A | 1.58e-20 | 24 | 329 | 21 | 339 | Thecrystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 [Bacillus sp. (in: Bacteria)],1V5D_A The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)],1V5D_B The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)] |
| 7CJU_A | 1.73e-20 | 24 | 329 | 27 | 345 | Crystalstructure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7CJU_B Crystal structure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7XGQ_A Chain A, chitosanase [Bacillus sp. K17-2],7XGQ_B Chain B, chitosanase [Bacillus sp. K17-2] |
| 6VC5_A | 3.06e-15 | 56 | 279 | 22 | 231 | 1.6Angstrom Resolution Crystal Structure of endoglucanase from Komagataeibacter sucrofermentans [Komagataeibacter sucrofermentans] |
| 1WZZ_A | 1.49e-09 | 69 | 276 | 48 | 243 | Structureof endo-beta-1,4-glucanase CMCax from Acetobacter xylinum [Komagataeibacter xylinus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19254 | 5.27e-27 | 27 | 325 | 58 | 359 | Beta-glucanase OS=Niallia circulans OX=1397 GN=bgc PE=3 SV=1 |
| P29019 | 8.66e-22 | 24 | 335 | 77 | 401 | Endoglucanase OS=Bacillus sp. (strain KSM-330) OX=72575 PE=1 SV=1 |
| P37696 | 4.47e-11 | 69 | 276 | 56 | 251 | Probable endoglucanase OS=Komagataeibacter hansenii OX=436 GN=cmcAX PE=1 SV=1 |
| P37651 | 2.24e-08 | 41 | 246 | 28 | 222 | Endoglucanase OS=Escherichia coli (strain K12) OX=83333 GN=bcsZ PE=1 SV=1 |
| Q8X5L9 | 5.31e-08 | 41 | 246 | 28 | 222 | Endoglucanase OS=Escherichia coli O157:H7 OX=83334 GN=bcsZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
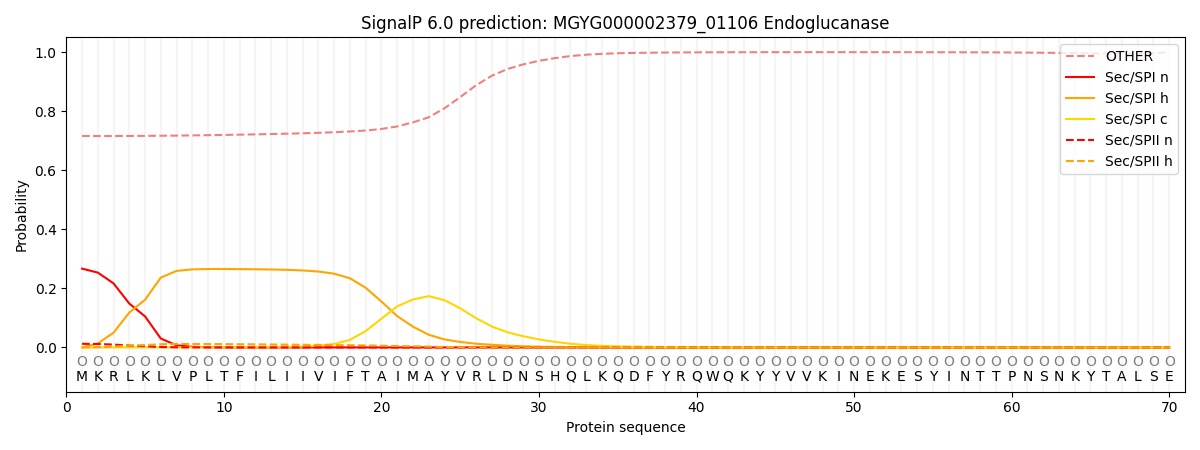
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.729674 | 0.248555 | 0.013892 | 0.000888 | 0.000542 | 0.006451 |