You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002386_01494
You are here: Home > Sequence: MGYG000002386_01494
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lactiplantibacillus plantarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactiplantibacillus; Lactiplantibacillus plantarum | |||||||||||
| CAZyme ID | MGYG000002386_01494 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1520257; End: 1521747 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 5.32e-25 | 394 | 488 | 3 | 96 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| COG0791 | Spr | 5.08e-19 | 394 | 480 | 89 | 179 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| PRK06347 | PRK06347 | 8.43e-14 | 23 | 143 | 472 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| pfam01476 | LysM | 1.93e-13 | 102 | 144 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| PRK06347 | PRK06347 | 4.36e-13 | 23 | 143 | 398 | 523 | 1,4-beta-N-acetylmuramoylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMX10699.1 | 2.11e-172 | 1 | 496 | 1 | 496 |
| AYA96693.1 | 2.11e-172 | 1 | 496 | 1 | 496 |
| ADN99001.1 | 2.11e-172 | 1 | 496 | 1 | 496 |
| ATL79173.1 | 2.11e-172 | 1 | 496 | 1 | 496 |
| QDX84757.1 | 2.11e-172 | 1 | 496 | 1 | 496 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6B8C_A | 3.21e-25 | 379 | 492 | 24 | 139 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
| 4Q4T_A | 2.40e-10 | 388 | 491 | 351 | 465 | Structureof the Resuscitation Promoting Factor Interacting protein RipA mutated at E444 [Mycobacterium tuberculosis H37Rv] |
| 2K1G_A | 2.48e-10 | 393 | 473 | 19 | 99 | SolutionNMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 [Escherichia coli K-12] |
| 2HBW_A | 9.21e-08 | 395 | 476 | 115 | 195 | ChainA, NLP/P60 protein [Trichormus variabilis ATCC 29413] |
| 3I86_A | 4.63e-07 | 393 | 491 | 25 | 134 | Crystalstructure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 [Mycobacterium avium subsp. paratuberculosis],3I86_B Crystal structure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 [Mycobacterium avium subsp. paratuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P13692 | 2.30e-22 | 379 | 492 | 398 | 513 | Protein P54 OS=Enterococcus faecium OX=1352 PE=3 SV=2 |
| P54421 | 1.38e-16 | 10 | 148 | 6 | 134 | Probable peptidoglycan endopeptidase LytE OS=Bacillus subtilis (strain 168) OX=224308 GN=lytE PE=1 SV=1 |
| A2RHZ5 | 1.80e-15 | 34 | 154 | 246 | 373 | Probable N-acetylmuramidase OS=Lactococcus lactis subsp. cremoris (strain MG1363) OX=416870 GN=acmA PE=3 SV=1 |
| P0C2T5 | 4.26e-15 | 34 | 154 | 246 | 373 | Probable N-acetylmuramidase OS=Lactococcus lactis subsp. cremoris OX=1359 GN=acmA PE=3 SV=1 |
| O34669 | 2.25e-14 | 25 | 154 | 20 | 132 | Cell wall-binding protein YocH OS=Bacillus subtilis (strain 168) OX=224308 GN=yocH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
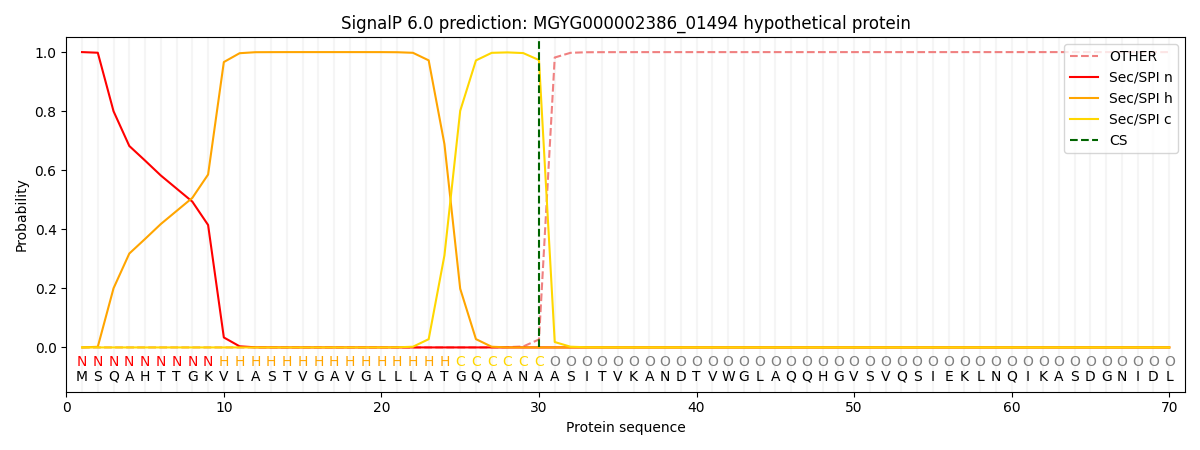
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000333 | 0.998930 | 0.000174 | 0.000203 | 0.000188 | 0.000164 |
