You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002449_03773
You are here: Home > Sequence: MGYG000002449_03773
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides acidifaciens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides acidifaciens | |||||||||||
| CAZyme ID | MGYG000002449_03773 | |||||||||||
| CAZy Family | GH50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 521; End: 2608 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH50 | 152 | 672 | 5.1e-108 | 0.8422664624808576 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1874 | GanA | 9.99e-05 | 391 | 527 | 119 | 251 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| cd19608 | GH113_mannanase-like | 3.65e-04 | 551 | 647 | 181 | 297 | Glycoside hydrolase family 113 beta-1,4-mannanase and similar proteins. Mannan endo-1,4-beta mannosidase (E.C 3.2.1.78) randomly cleaves (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans and is also called beta-1,4-mannanase, endo-1,4-beta-mannanase, endo-beta-1,4-mannase, beta-mannanase B, beta-1, 4-mannan 4-mannanohydrolase, endo-beta-mannanase, beta-D-mannanase, 1,4-beta-D-mannan mannanohydrolase, and 4-beta-D-mannan mannanohydrolase. (1->4)-beta-linked mannans are polysaccharides with a linear polymer backbone of (1->4)-beta-linked mannose units (in plants and fungi) or alternating mannose and glucose/galactose units (glucomannan in plants and fungi, and galactomannan and galactoglucomannan in plants), such as in the hemicellulose fraction of hard- and softwoods. Complete degradation of mannan requires a series of enzymes, including beta-1,4-mannanase. According to the CAZy database beta-1,4-mannanases are grouped into various glycoside hydrolase (GH) families; GH family 113 beta-1,4-mannanases include mostly bacterial and archaeal sequences. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGY44025.1 | 4.41e-220 | 4 | 692 | 6 | 704 |
| AFN74211.1 | 3.28e-204 | 6 | 692 | 11 | 716 |
| SQI63245.1 | 9.33e-153 | 73 | 692 | 29 | 642 |
| QHW33563.1 | 2.90e-148 | 74 | 681 | 52 | 665 |
| AIQ37144.1 | 2.92e-141 | 75 | 682 | 51 | 653 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XJ9_A | 9.47e-61 | 50 | 673 | 55 | 760 | Structureof PfGH50B [Pseudoalteromonas fuliginea],6XJ9_B Structure of PfGH50B [Pseudoalteromonas fuliginea] |
| 4BQ2_A | 3.33e-53 | 203 | 679 | 204 | 750 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4BQ4_A | 8.44e-53 | 203 | 679 | 204 | 750 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ4_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 5Z6P_A | 2.94e-51 | 203 | 672 | 223 | 759 | Thecrystal structure of an agarase, AgWH50C [Agarivorans gilvus],5Z6P_B The crystal structure of an agarase, AgWH50C [Agarivorans gilvus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48840 | 1.61e-58 | 53 | 679 | 268 | 955 | Beta-agarase B OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaB PE=3 SV=1 |
| P48839 | 1.92e-31 | 266 | 672 | 467 | 913 | Beta-agarase A OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
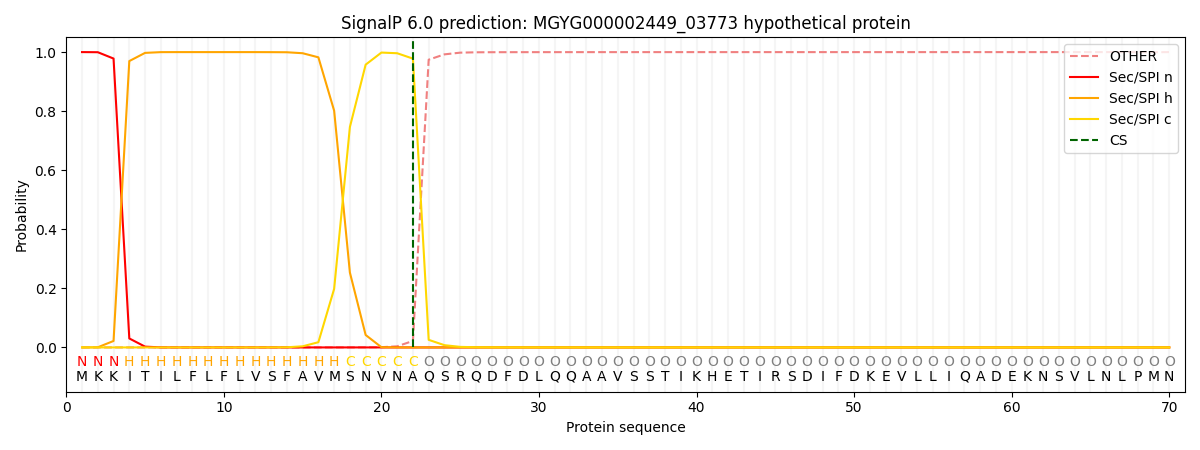
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000277 | 0.998997 | 0.000212 | 0.000173 | 0.000165 | 0.000148 |
