You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002561_01550
You are here: Home > Sequence: MGYG000002561_01550
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
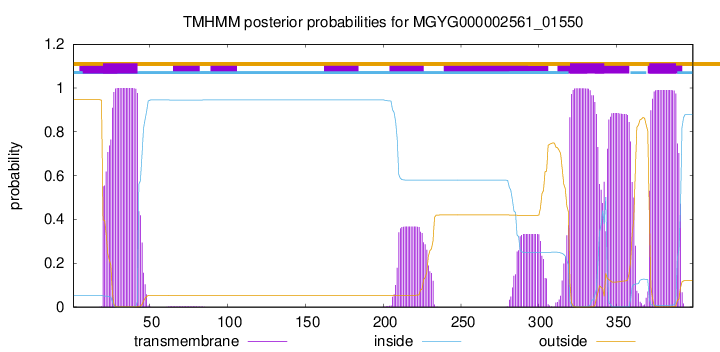
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902388495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902388495 | |||||||||||
| CAZyme ID | MGYG000002561_01550 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5471; End: 6670 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 62 | 282 | 1.6e-33 | 0.9782608695652174 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13641 | Glyco_tranf_2_3 | 3.91e-43 | 62 | 282 | 3 | 228 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
| cd06438 | EpsO_like | 3.34e-14 | 84 | 223 | 19 | 162 | EpsO protein participates in the methanolan synthesis. The Methylobacillus sp EpsO protein is predicted to participate in the methanolan synthesis. Methanolan is an exopolysaccharide (EPS), composed of glucose, mannose and galactose. A 21 genes cluster was predicted to participate in the methanolan synthesis. Gene disruption analysis revealed that EpsO is one of the glycosyltransferase enzymes involved in the synthesis of repeating sugar units onto the lipid carrier. |
| COG1215 | BcsA | 2.21e-08 | 13 | 391 | 5 | 387 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMI81774.1 | 1.26e-244 | 1 | 398 | 1 | 398 |
| QUT68139.1 | 1.80e-244 | 1 | 398 | 1 | 398 |
| QQA31515.1 | 1.80e-244 | 1 | 398 | 1 | 398 |
| QUT63158.1 | 1.80e-244 | 1 | 398 | 1 | 398 |
| QUT36865.1 | 1.80e-244 | 1 | 398 | 1 | 398 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
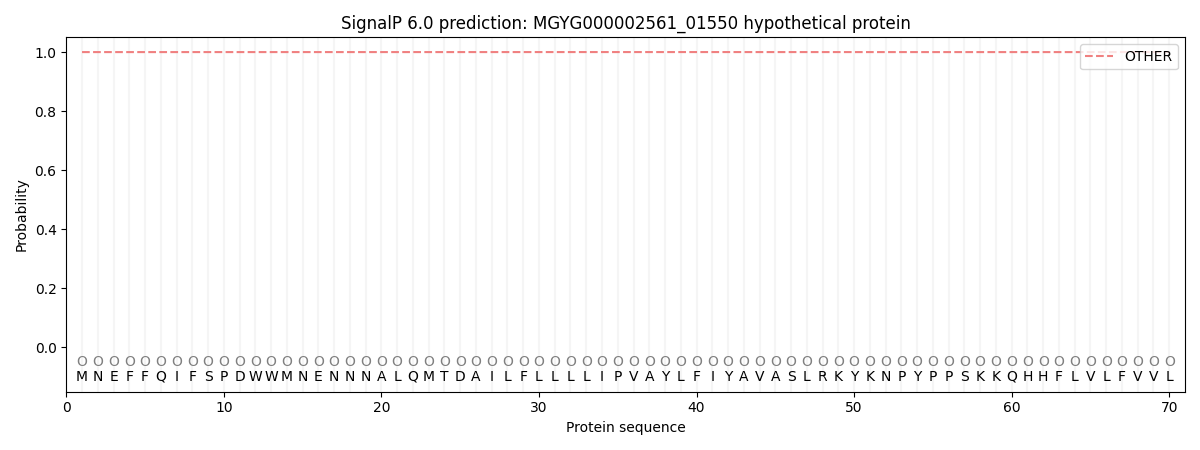
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000066 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |