You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002657_02370
You are here: Home > Sequence: MGYG000002657_02370
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
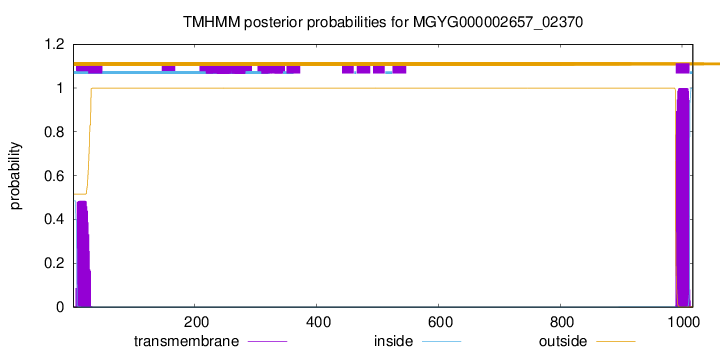
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-95; | |||||||||||
| CAZyme ID | MGYG000002657_02370 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4486; End: 7542 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.02e-14 | 224 | 467 | 13 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| NF033839 | PspC_subgroup_2 | 0.005 | 740 | 803 | 445 | 502 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| COG3147 | DedD | 0.007 | 745 | 810 | 87 | 152 | Cell division protein DedD (periplasmic protein involved in septation) [Cell cycle control, cell division, chromosome partitioning]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM02197.1 | 3.02e-106 | 77 | 477 | 43 | 451 |
| ACR71571.1 | 2.64e-60 | 106 | 477 | 60 | 438 |
| CCO04280.1 | 3.50e-59 | 113 | 477 | 54 | 424 |
| AEN96074.1 | 8.85e-59 | 97 | 473 | 144 | 518 |
| CBK91761.1 | 2.39e-58 | 81 | 473 | 130 | 533 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
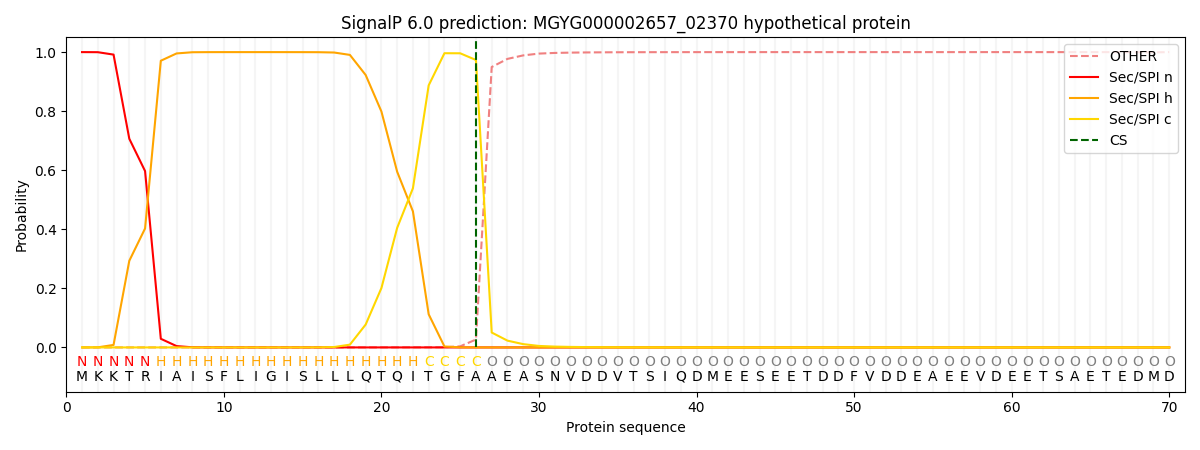
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000916 | 0.998049 | 0.000371 | 0.000241 | 0.000209 | 0.000186 |