You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002836_00784
You are here: Home > Sequence: MGYG000002836_00784
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
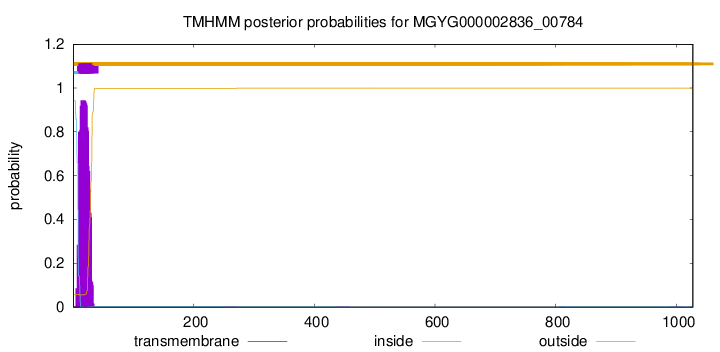
TMHMM annotations
Basic Information help
| Species | Anaerocolumna aminovalerica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerocolumna; Anaerocolumna aminovalerica | |||||||||||
| CAZyme ID | MGYG000002836_00784 | |||||||||||
| CAZy Family | CBM3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 74516; End: 77599 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 5.61e-17 | 686 | 924 | 45 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 2.64e-12 | 317 | 375 | 1 | 54 | Bacterial SH3 domain. |
| pfam08239 | SH3_3 | 7.94e-09 | 132 | 188 | 3 | 54 | Bacterial SH3 domain. |
| COG3103 | YgiM | 7.76e-08 | 1 | 189 | 1 | 151 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| smart00287 | SH3b | 1.15e-05 | 314 | 372 | 6 | 59 | Bacterial SH3 domain homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ93456.1 | 2.21e-282 | 1 | 1026 | 1 | 895 |
| QHQ61172.1 | 1.97e-263 | 1 | 1027 | 1 | 768 |
| ABX43817.1 | 6.04e-232 | 35 | 1026 | 194 | 1279 |
| BCJ97942.1 | 4.94e-228 | 1 | 1026 | 1 | 907 |
| BCN29841.1 | 1.68e-219 | 14 | 1027 | 11 | 890 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O32041 | 7.05e-08 | 32 | 187 | 97 | 239 | Putative N-acetylmuramoyl-L-alanine amidase YrvJ OS=Bacillus subtilis (strain 168) OX=224308 GN=yrvJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
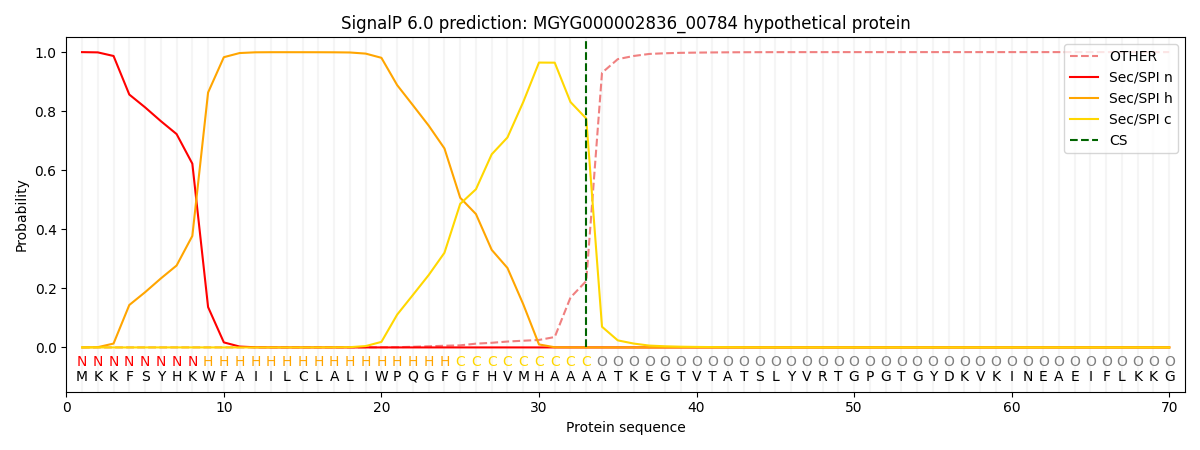
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000476 | 0.998754 | 0.000227 | 0.000193 | 0.000163 | 0.000159 |