You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002963_02888
You are here: Home > Sequence: MGYG000002963_02888
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaerosporobacter mobilis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerosporobacter; Anaerosporobacter mobilis | |||||||||||
| CAZyme ID | MGYG000002963_02888 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9143; End: 12961 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 281 | 568 | 2.8e-72 | 0.46621621621621623 |
| GH128 | 730 | 973 | 3.4e-54 | 0.9732142857142857 |
| CBM32 | 1016 | 1128 | 2.3e-30 | 0.9193548387096774 |
| CBM32 | 1157 | 1270 | 1e-29 | 0.9193548387096774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11790 | Glyco_hydro_cc | 1.24e-49 | 724 | 973 | 8 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| pfam00754 | F5_F8_type_C | 6.86e-24 | 1013 | 1126 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 6.87e-22 | 1154 | 1268 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.80e-09 | 1143 | 1272 | 1 | 142 | Substituted updates: Jan 31, 2002 |
| cd00057 | FA58C | 6.80e-09 | 1016 | 1115 | 16 | 130 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADQ45027.1 | 7.11e-232 | 42 | 1131 | 1473 | 2433 |
| CUU48277.1 | 1.14e-198 | 44 | 598 | 195 | 752 |
| AVK46774.1 | 6.30e-198 | 44 | 598 | 195 | 752 |
| QUN33032.1 | 1.25e-197 | 44 | 598 | 195 | 752 |
| QUF71865.1 | 1.25e-197 | 44 | 598 | 195 | 752 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4TZ1_A | 9.38e-150 | 44 | 584 | 9 | 533 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4TZ3_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4TZ5_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4TZ5_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
| 4TYV_A | 1.00e-149 | 44 | 584 | 11 | 535 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4TYV_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E] |
| 4PEX_A | 1.39e-149 | 44 | 584 | 21 | 545 | Structureof the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEX_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEY_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4PEZ_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4PF0_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4PF0_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
| 4PEW_A | 1.05e-148 | 44 | 584 | 21 | 545 | Structureof sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4PEW_B Structure of sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 2RV9_A | 2.00e-33 | 1007 | 1131 | 7 | 136 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2NFJ9 | 4.13e-149 | 44 | 584 | 65 | 589 | Exo-beta-1,3-glucanase OS=Streptomyces sp. (strain SirexAA-E / ActE) OX=862751 GN=SACTE_4363 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
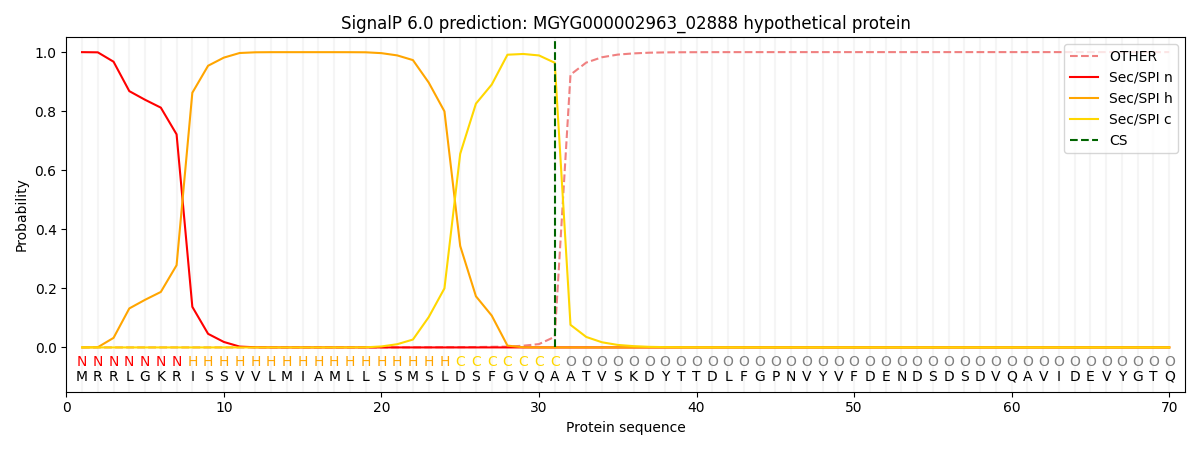
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000273 | 0.998774 | 0.000413 | 0.000204 | 0.000167 | 0.000139 |
