You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003132_01462
You are here: Home > Sequence: MGYG000003132_01462
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Dysgonomonas sp900556485 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas sp900556485 | |||||||||||
| CAZyme ID | MGYG000003132_01462 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 81298; End: 83094 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 88 | 350 | 6.4e-17 | 0.7777777777777778 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06439 | DUF1080 | 6.40e-57 | 385 | 595 | 1 | 182 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
| cd15482 | Sialidase_non-viral | 1.20e-21 | 87 | 371 | 49 | 338 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13088 | BNR_2 | 9.01e-14 | 92 | 349 | 27 | 278 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam13088 | BNR_2 | 3.03e-09 | 46 | 269 | 35 | 257 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| cd15482 | Sialidase_non-viral | 6.31e-06 | 179 | 309 | 27 | 178 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDU73633.1 | 4.10e-117 | 38 | 392 | 208 | 560 |
| QDV35524.1 | 7.83e-83 | 17 | 375 | 17 | 377 |
| AQW27075.1 | 1.47e-55 | 38 | 382 | 203 | 569 |
| ATD48363.1 | 1.47e-55 | 38 | 382 | 203 | 569 |
| ABG82852.1 | 3.89e-55 | 38 | 382 | 203 | 569 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3OSD_A | 1.44e-87 | 386 | 597 | 38 | 264 | Crystalstructure of a putative glycosyl hydrolase (BT2157) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.80 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 3NMB_A | 1.48e-85 | 382 | 597 | 29 | 259 | Crystalstructure of a putative sugar hydrolase (BACOVA_03189) from Bacteroides ovatus at 2.40 A resolution [Bacteroides ovatus ATCC 8483] |
| 4HXC_A | 3.05e-82 | 386 | 597 | 36 | 262 | Crystalstructure of a putative glycosyl hydrolase (BACUNI_00951) from Bacteroides uniformis ATCC 8492 at 2.15 A resolution [Bacteroides uniformis ATCC 8492] |
| 3H3L_A | 3.01e-64 | 385 | 597 | 22 | 241 | Crystalstructure of PUTATIVE SUGAR HYDROLASE (YP_001304206.1) from Parabacteroides distasonis ATCC 8503 at 1.59 A resolution [Parabacteroides distasonis ATCC 8503],3H3L_B Crystal structure of PUTATIVE SUGAR HYDROLASE (YP_001304206.1) from Parabacteroides distasonis ATCC 8503 at 1.59 A resolution [Parabacteroides distasonis ATCC 8503],3H3L_C Crystal structure of PUTATIVE SUGAR HYDROLASE (YP_001304206.1) from Parabacteroides distasonis ATCC 8503 at 1.59 A resolution [Parabacteroides distasonis ATCC 8503] |
| 4JQT_A | 2.62e-50 | 386 | 597 | 223 | 430 | Crystalstructure of a putative glycosyl hydrolase (BT3469) from Bacteroides thetaiotaomicron VPI-5482 at 2.49 A resolution [Bacteroides thetaiotaomicron VPI-5482],4JQT_B Crystal structure of a putative glycosyl hydrolase (BT3469) from Bacteroides thetaiotaomicron VPI-5482 at 2.49 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
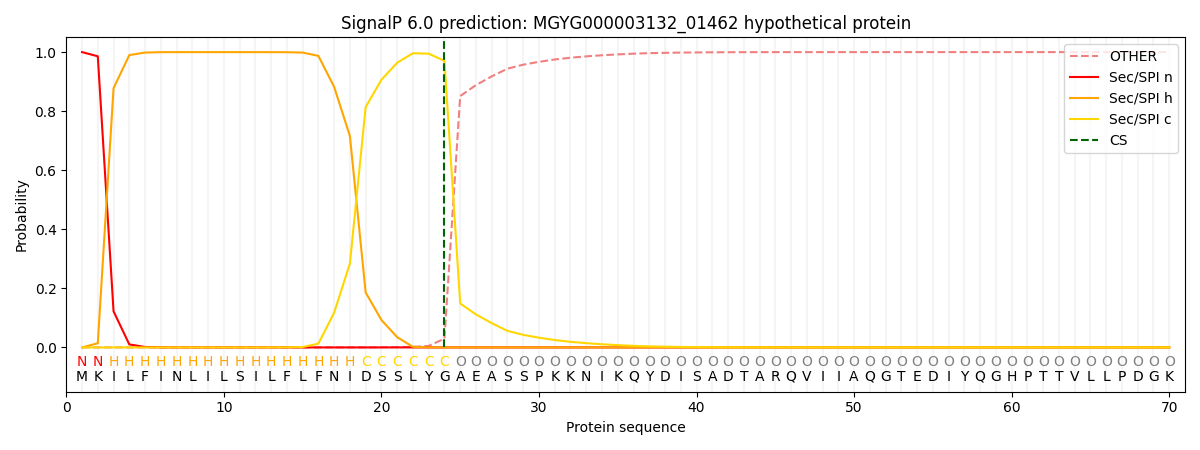
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000678 | 0.998444 | 0.000361 | 0.000179 | 0.000162 | 0.000167 |
