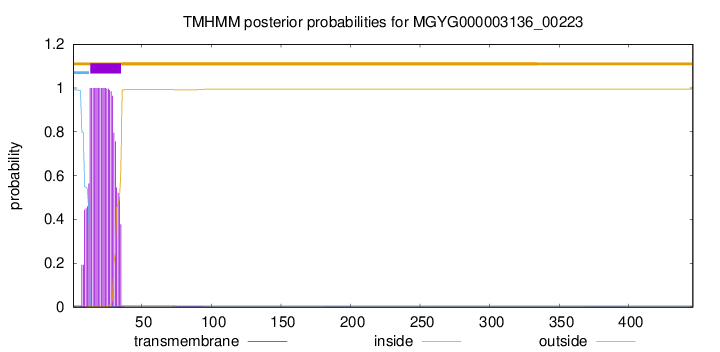
You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003136_00223
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus;
|
| CAZyme ID |
MGYG000003136_00223
|
| CAZy Family |
GH73 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003136 |
1781350 |
MAG |
United States |
North America |
|
| Gene Location |
Start: 11859;
End: 13199
Strand: -
|
No EC number prediction in MGYG000003136_00223.
| Family |
Start |
End |
Evalue |
family coverage |
| GH73 |
72 |
216 |
8.7e-16 |
0.953125 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1705
|
FlgJ |
5.04e-12 |
17 |
222 |
4 |
188 |
Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
This protein is predicted as OTHER
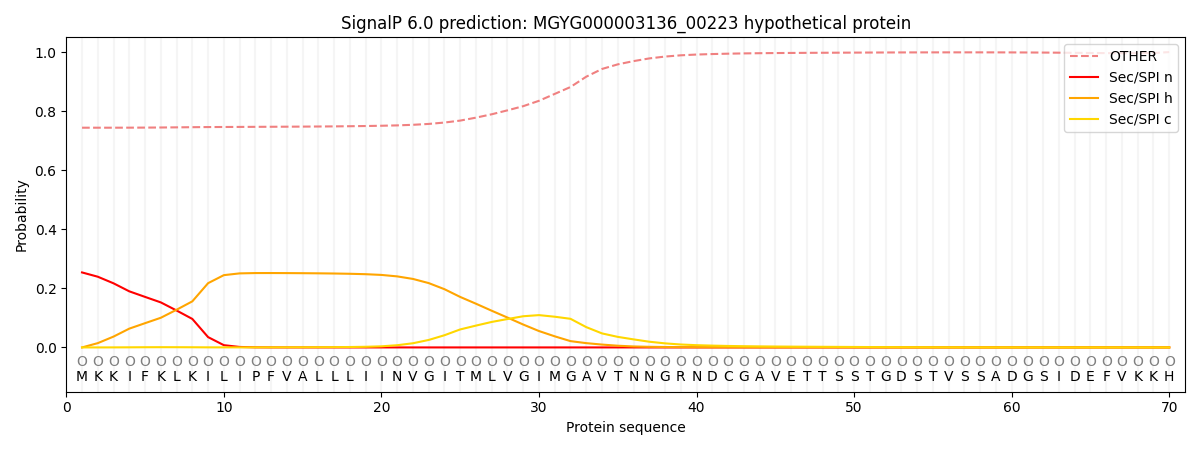
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.757226
|
0.237502
|
0.001047
|
0.000812
|
0.000449
|
0.002968
|