You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003199_00551
You are here: Home > Sequence: MGYG000003199_00551
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | HGM05190 sp900759815 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; HGM05190; HGM05190 sp900759815 | |||||||||||
| CAZyme ID | MGYG000003199_00551 | |||||||||||
| CAZy Family | PL4 | |||||||||||
| CAZyme Description | Rhamnogalacturonate lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4561; End: 6480 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL4 | 33 | 571 | 1.6e-116 | 0.9836660617059891 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10320 | RGL4_N | 8.00e-32 | 37 | 284 | 12 | 259 | N-terminal catalytic domain of rhamnogalacturonan lyase, a family 4 polysaccharide lyase. The rhamnogalacturonan lyase of the polysaccharide lyase family 4 (RGL4) is involved in the degradation of RG (rhamnogalacturonan) type-I, an important pectic plant cell wall polysaccharide, by cleaving the alpha-1,4 glycoside bond between L-rhamnose and D-galacturonic acids in the backbone of RG type-I through a beta-elimination reaction. RGL4 consists of three domains, an N-terminal catalytic domain, a middle domain with a FNIII type fold and a C-terminal domain with a jelly roll fold; the middle and C-terminal domains are both putative carbohydrate binding modules. There are two types of RG lyases, which both cleave the alpha-1,4 bonds of the RG-I main chain (RG chain) through the beta-elimination reaction, but belong to two structurally unrelated polysaccharide lyase (PL) families, 4 and 11. |
| cd10317 | RGL4_C | 4.40e-28 | 424 | 574 | 4 | 161 | C-terminal domain of rhamnogalacturonan lyase, a family 4 polysaccharide lyase. The rhamnogalacturonan lyase of the polysaccharide lyase family 4 (RGL4) is involved in the degradation of RG (rhamnogalacturonan) type-I, an important pectic plant cell wall polysaccharide, by cleaving the alpha-1,4 glycoside bond between L-rhamnose and D-galacturonic acids in the backbone of RG type-I through a beta-elimination reaction. RGL4 consists of three domains, an N-terminal catalytic domain, a middle domain with a FNIII type fold and a C-terminal domain with a jelly roll fold. Both the middle and the C-terminal domain are putative carbohydrate binding modules. There are two types of RG lyases, which both cleave the alpha-1,4 bonds of the RG-I main chain (RG chain) through the beta-elimination reaction, but belong to two structurally unrelated polysaccharide lyase (PL) families, 4 and 11. |
| pfam14683 | CBM-like | 2.56e-23 | 423 | 573 | 5 | 157 | Polysaccharide lyase family 4, domain III. CBM-like is domain III of rhamnogalacturonan lyase (RG-lyase). The full-length protein specifically recognizes and cleaves alpha-1,4 glycosidic bonds between l-rhamnose and d-galacturonic acids in the backbone of rhamnogalacturonan-I, a major component of the plant cell wall polysaccharide, pectin. This domain possesses a jelly roll beta-sandwich fold structurally homologous to carbohydrate binding modules (CBMs), and it carries two sulfate ions and a hexa-coordinated calcium ion. |
| cd10316 | RGL4_M | 4.22e-09 | 318 | 399 | 1 | 81 | Middle domain of rhamnogalacturonan lyase, a family 4 polysaccharide lyase. The rhamnogalacturonan lyase of the polysaccharide lyase family 4 (RGL4) is involved in the degradation of RG (rhamnogalacturonan) type-I, an important pectic plant cell wall polysaccharide, by cleaving the alpha-1,4 glycoside bond between L-rhamnose and D-galacturonic acids in the backbone of RG type-I through a beta-elimination reaction. RGL4 consists of three domains, an N-terminal catalytic domain, a middle domain with a FNIII type fold and a C-terminal domain with a jelly roll fold. Both the middle domain represented by this model and the C-terminal domain are putative carbohydrate binding modules. There are two types of RG lyases, which both cleave the alpha-1,4 bonds of the RG-I main chain (RG chain) through the beta-elimination reaction, but belong to two structurally unrelated polysaccharide lyase (PL) families, 4 and 11. |
| pfam14686 | fn3_3 | 8.63e-05 | 339 | 386 | 9 | 56 | Polysaccharide lyase family 4, domain II. FnIII-like is domain II of rhamnogalacturonan lyase (RG-lyase). The full-length protein specifically recognizes and cleaves alpha-1,4 glycosidic bonds between l-rhamnose and d-galacturonic acids in the backbone of rhamnogalacturonan-I, a major component of the plant cell wall polysaccharide, pectin. This domain displays an immunoglobulin-like or more specifically Fibronectin-III type fold and shows highest structural similarity to the C-terminal beta-sandwich subdomain of the pro-hormone/propeptide processing enzyme carboxypeptidase gp180 from duck. It serves to assist in producing the deep pocket, with domain III, into which the substrate fits. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBH93641.1 | 4.12e-110 | 20 | 579 | 42 | 585 |
| BAV08848.1 | 1.86e-95 | 74 | 574 | 96 | 594 |
| QOH51094.1 | 9.47e-82 | 37 | 573 | 38 | 571 |
| QOH46789.1 | 9.47e-82 | 37 | 573 | 38 | 571 |
| ATZ93356.1 | 9.47e-82 | 37 | 573 | 38 | 571 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8RJP2 | 2.28e-79 | 37 | 573 | 40 | 573 | Rhamnogalacturonate lyase OS=Dickeya dadantii (strain 3937) OX=198628 GN=rhiE PE=1 SV=1 |
| Q2U5P7 | 2.37e-17 | 66 | 510 | 68 | 558 | Probable rhamnogalacturonate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=rglC PE=3 SV=1 |
| Q5AZ85 | 3.14e-14 | 74 | 464 | 81 | 503 | Rhamnogalacturonate lyase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=rglB PE=2 SV=2 |
| B0XPA2 | 1.29e-10 | 74 | 464 | 82 | 501 | Probable rhamnogalacturonate lyase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=rglB PE=3 SV=1 |
| Q4WR79 | 1.29e-10 | 74 | 464 | 82 | 501 | Probable rhamnogalacturonate lyase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=rglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
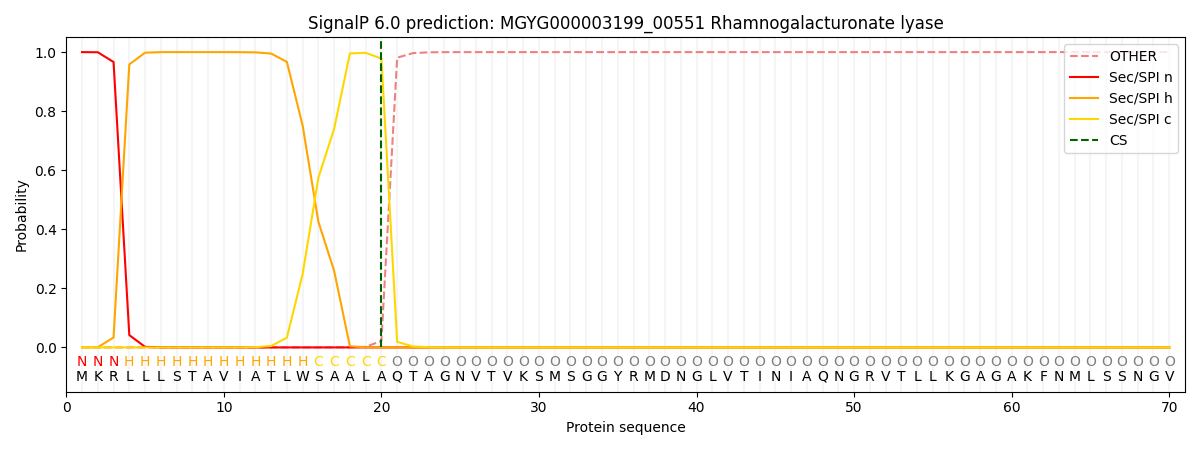
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000245 | 0.999045 | 0.000184 | 0.000172 | 0.000161 | 0.000149 |
