You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003199_01315
You are here: Home > Sequence: MGYG000003199_01315
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
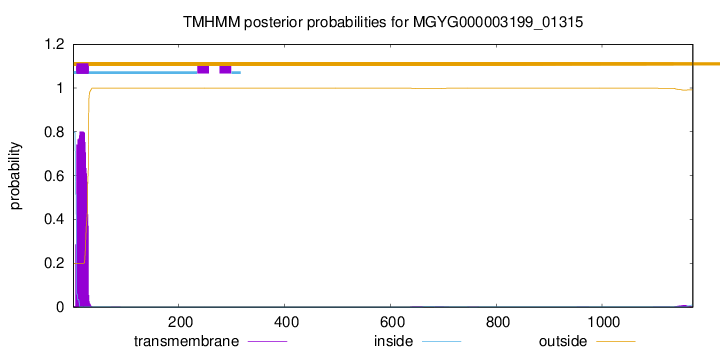
TMHMM annotations
Basic Information help
| Species | HGM05190 sp900759815 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; HGM05190; HGM05190 sp900759815 | |||||||||||
| CAZyme ID | MGYG000003199_01315 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4160; End: 7678 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 78 | 258 | 5e-52 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00063 | FN3 | 4.87e-10 | 466 | 553 | 3 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| COG3866 | PelB | 2.14e-09 | 13 | 290 | 14 | 304 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00060 | FN3 | 8.21e-08 | 467 | 540 | 4 | 81 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| smart00656 | Amb_all | 5.24e-07 | 76 | 262 | 1 | 190 | Amb_all domain. |
| pfam00041 | fn3 | 1.18e-04 | 466 | 540 | 2 | 80 | Fibronectin type III domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCP72135.1 | 0.0 | 23 | 1171 | 23 | 1185 |
| QCD38445.1 | 2.05e-284 | 23 | 644 | 23 | 652 |
| QUT73817.1 | 1.22e-184 | 26 | 683 | 22 | 689 |
| QUT74519.1 | 1.23e-160 | 14 | 653 | 12 | 636 |
| QUT90499.1 | 3.54e-150 | 19 | 459 | 29 | 464 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B0XMA2 | 2.23e-123 | 13 | 451 | 7 | 408 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
| Q4WL88 | 2.23e-123 | 13 | 451 | 7 | 408 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
| A1DPF0 | 3.11e-123 | 13 | 451 | 7 | 408 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
| B8NQQ7 | 2.34e-121 | 27 | 451 | 20 | 407 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q5B297 | 6.81e-121 | 27 | 466 | 20 | 420 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
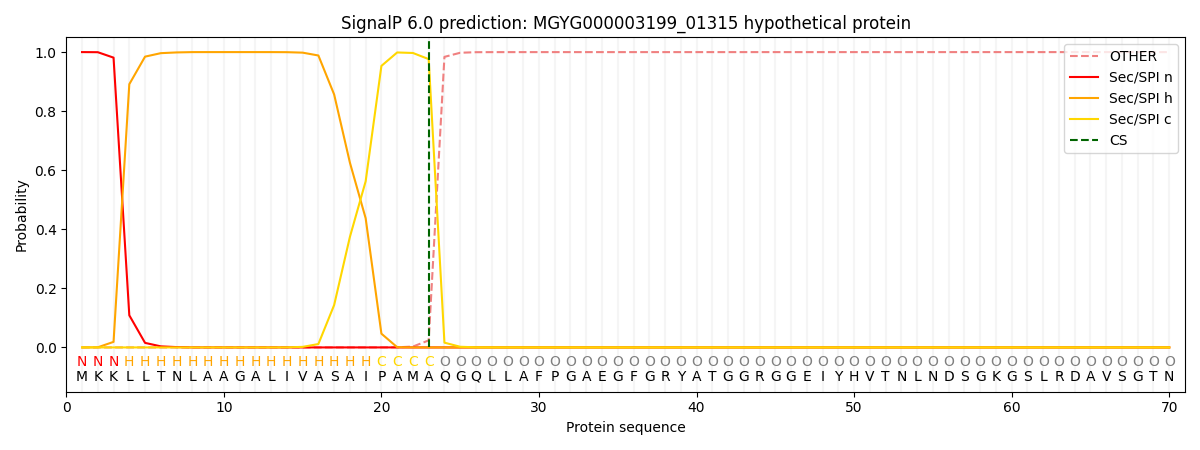
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000241 | 0.999089 | 0.000181 | 0.000171 | 0.000160 | 0.000143 |