You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003282_00598
You are here: Home > Sequence: MGYG000003282_00598
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprobacter sp900545915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter sp900545915 | |||||||||||
| CAZyme ID | MGYG000003282_00598 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 113479; End: 115452 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 17 | 387 | 4.4e-117 | 0.9893333333333333 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 9.86e-10 | 105 | 288 | 2 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| TIGR04183 | Por_Secre_tail | 4.75e-06 | 600 | 656 | 13 | 70 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| pfam18962 | Por_Secre_tail | 1.19e-05 | 600 | 656 | 11 | 71 | Secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber have on average twenty or more copies of this C-terminal domain, associated with sorting to the outer membrane and covalent modification. This domain targets proteins to type IX secretion systems and is secreted then cleaved off by a C-terminal signal peptidease. Based on similarity to other families it is likely that this domain adopts an immunoglobulin like fold. |
| NF033708 | T9SS_Cterm_ChiA | 1.92e-05 | 608 | 657 | 2 | 55 | T9SS sorting signal type C. The sorting signals of type IX secretion systems (T9SS) in the CFB bacteria are long, compared to other prokaryotic C-terminal sorting motif-containing signals, including LPXTG, PEP-CTERM, and GlyGly-TERM, and they seem to contain multiple motifs. A few T9SS substrates, including ChiA, have a variant form of T9SS sorting signal that may score poorly to both TIGR04183 (type A) and TIGR04131 (type B), depend on T9SS for secretion, but are released from the cell rather than left anchored to the cell surface. |
| cd14251 | PL-6 | 7.52e-04 | 42 | 143 | 8 | 106 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APZ45418.1 | 1.02e-113 | 41 | 410 | 224 | 583 |
| AUC19279.1 | 1.02e-113 | 41 | 410 | 224 | 583 |
| QQY82139.1 | 2.10e-112 | 32 | 410 | 27 | 395 |
| AOW17787.1 | 1.26e-109 | 38 | 410 | 216 | 578 |
| APU68883.1 | 1.91e-108 | 16 | 411 | 232 | 616 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 1.61e-74 | 21 | 410 | 17 | 398 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 2.17e-33 | 20 | 375 | 4 | 407 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 1.60e-75 | 21 | 410 | 42 | 423 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 1.77e-73 | 21 | 410 | 42 | 423 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P22751 | 3.23e-24 | 18 | 299 | 388 | 646 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
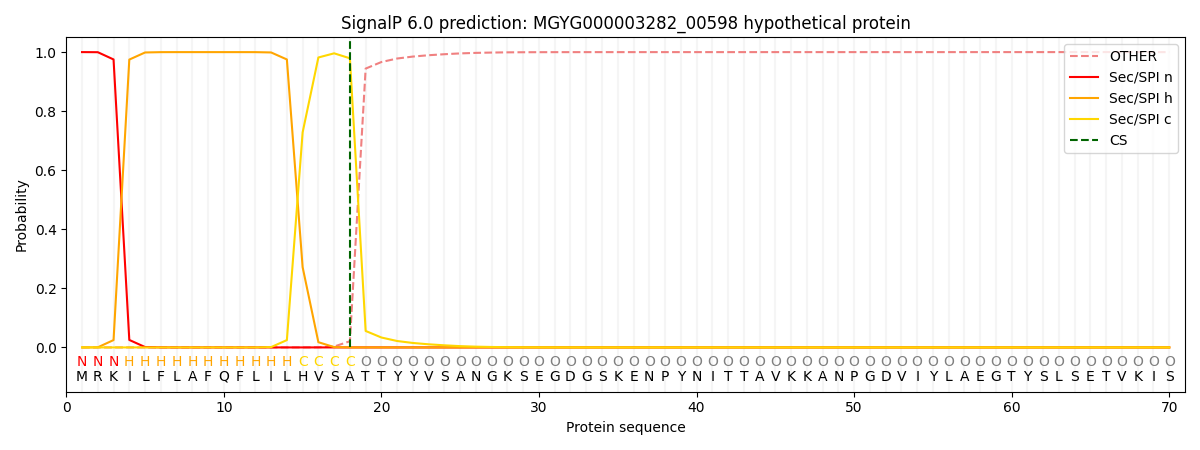
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000335 | 0.999022 | 0.000173 | 0.000154 | 0.000148 | 0.000144 |
