You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003362_00244
You are here: Home > Sequence: MGYG000003362_00244
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
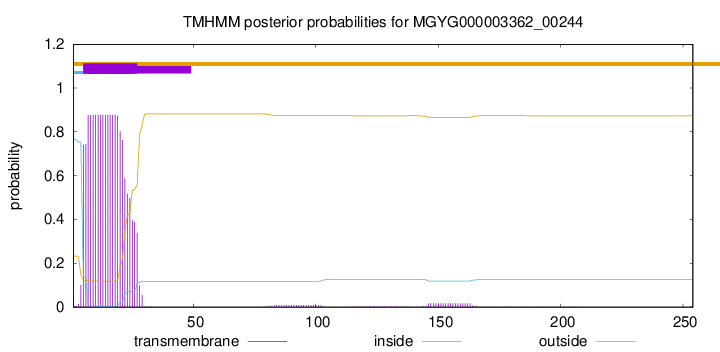
TMHMM annotations
Basic Information help
| Species | Dysgonomonas sp900079735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas sp900079735 | |||||||||||
| CAZyme ID | MGYG000003362_00244 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 279925; End: 280689 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 31 | 234 | 1.8e-31 | 0.8898678414096917 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 5.27e-48 | 30 | 253 | 171 | 386 | Predicted peptidase [General function prediction only]. |
| COG1506 | DAP2 | 5.43e-15 | 28 | 233 | 373 | 594 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam02230 | Abhydrolase_2 | 4.34e-13 | 45 | 236 | 10 | 202 | Phospholipase/Carboxylesterase. This family consists of both phospholipases and carboxylesterases with broad substrate specificity, and is structurally related to alpha/beta hydrolases pfam00561. |
| COG0400 | YpfH | 5.73e-13 | 45 | 230 | 14 | 186 | Predicted esterase [General function prediction only]. |
| COG3509 | LpqC | 9.85e-11 | 24 | 205 | 38 | 205 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABS60377.1 | 4.78e-49 | 20 | 254 | 2 | 245 |
| BCI61582.1 | 7.80e-45 | 31 | 250 | 823 | 1039 |
| QDU56037.1 | 7.61e-35 | 33 | 254 | 801 | 1007 |
| ACR12533.1 | 1.90e-32 | 32 | 251 | 63 | 278 |
| VTR91196.1 | 9.27e-32 | 33 | 254 | 43 | 239 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 6.04e-51 | 30 | 254 | 155 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 1.00e-22 | 30 | 254 | 18 | 217 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D4AR77 | 2.52e-09 | 20 | 233 | 375 | 605 | Uncharacterized secreted protein ARB_06907 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_06907 PE=1 SV=2 |
| Q7XR62 | 1.01e-08 | 52 | 234 | 14 | 202 | Probable inactive carboxylesterase Os04g0669700 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0669700 PE=3 SV=1 |
| Q4I8Q4 | 1.71e-08 | 46 | 235 | 15 | 215 | Acyl-protein thioesterase 1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=FGRAMPH1_01T20223 PE=3 SV=1 |
| F0U7H7 | 1.16e-07 | 29 | 249 | 646 | 885 | Probable dipeptidyl-aminopeptidase B OS=Ajellomyces capsulatus (strain H88) OX=544711 GN=DAPB PE=3 SV=1 |
| C6HRC7 | 1.16e-07 | 29 | 249 | 646 | 885 | Probable dipeptidyl-aminopeptidase B OS=Ajellomyces capsulatus (strain H143) OX=544712 GN=DAPB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
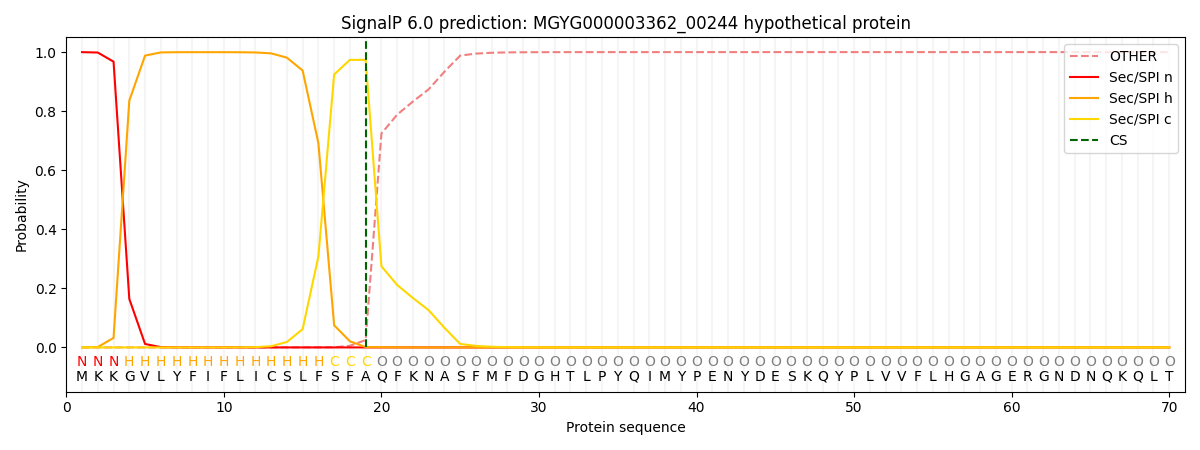
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001148 | 0.997555 | 0.000682 | 0.000195 | 0.000197 | 0.000196 |