You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003431_00739
You are here: Home > Sequence: MGYG000003431_00739
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; HUN007; | |||||||||||
| CAZyme ID | MGYG000003431_00739 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 369; End: 2534 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 48 | 503 | 5.7e-153 | 0.9824561403508771 |
| CBM62 | 522 | 657 | 4.3e-32 | 0.9770992366412213 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 1.02e-21 | 55 | 366 | 18 | 364 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 1.80e-15 | 157 | 506 | 124 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam00754 | F5_F8_type_C | 3.90e-07 | 525 | 600 | 1 | 79 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd14256 | Dockerin_I | 4.90e-06 | 667 | 704 | 2 | 41 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 1.49e-05 | 667 | 704 | 1 | 40 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU23821.1 | 1.61e-273 | 1 | 690 | 1 | 679 |
| QTE67528.1 | 7.42e-176 | 33 | 505 | 28 | 510 |
| CBK73885.1 | 1.81e-170 | 30 | 664 | 943 | 1646 |
| AOZ97766.1 | 8.59e-138 | 25 | 506 | 51 | 582 |
| QMW77599.1 | 1.94e-134 | 5 | 516 | 23 | 771 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FMV_A | 1.20e-12 | 157 | 504 | 89 | 385 | CrystalStructure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
| 2YB7_A | 4.32e-12 | 521 | 657 | 7 | 138 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YB7_B Chain B, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YFU_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YFZ_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YG0_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 4QAW_A | 8.35e-12 | 157 | 506 | 85 | 389 | Structureof modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_B Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_C Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_D Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_E Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_F Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_G Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_H Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 5G56_A | 4.41e-11 | 488 | 657 | 673 | 839 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 7O0E_A | 2.13e-08 | 53 | 521 | 17 | 445 | ChainA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464],7O0E_G Chain G, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q76FP5 | 5.65e-63 | 33 | 508 | 20 | 476 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
| A0A401ETL2 | 3.61e-34 | 5 | 516 | 6 | 621 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| Q6YK37 | 5.22e-10 | 157 | 505 | 118 | 421 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
| G2Q1N4 | 2.32e-08 | 53 | 521 | 41 | 469 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
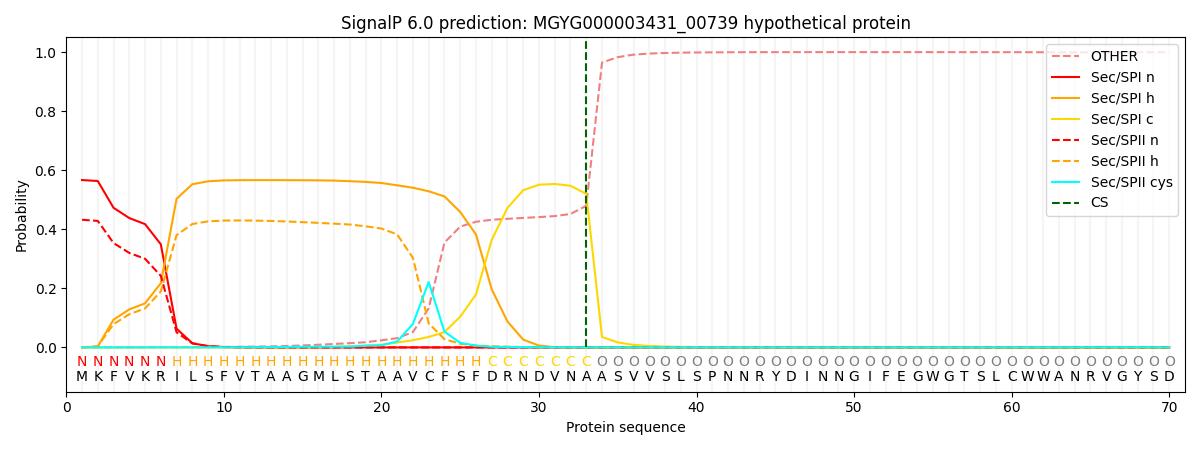
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001392 | 0.562189 | 0.435497 | 0.000533 | 0.000202 | 0.000152 |
