You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003447_01061
You are here: Home > Sequence: MGYG000003447_01061
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; | |||||||||||
| CAZyme ID | MGYG000003447_01061 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Glycosyl hydrolase family 109 protein 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 725; End: 2188 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 58 | 479 | 2.2e-157 | 0.9924812030075187 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.52e-18 | 62 | 414 | 4 | 330 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 4.32e-15 | 62 | 188 | 1 | 118 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| COG4091 | COG4091 | 0.003 | 62 | 158 | 18 | 128 | Predicted homoserine dehydrogenase, contains C-terminal SAF domain [Amino acid transport and metabolism]. |
| cd17541 | REC_CheB-like | 0.003 | 120 | 172 | 32 | 113 | phosphoacceptor receiver (REC) domain of chemotaxis response regulator protein-glutamate methylesterase CheB and similar chemotaxis proteins. Methylesterase CheB is a chemotaxis response regulator with an N-terminal REC domain and a C-terminal methylesterase domain. Chemotaxis is a behavior known in motile bacteria that directs their movement in response to chemical gradients. CheB is a phosphorylation-activated response regulator involved in the reversible modification of bacterial chemotaxis receptors. It catalyzes the demethylation of specific methylglutamate residues introduced into the chemoreceptors (methyl-accepting chemotaxis proteins) by CheR. The CheB REC domain packs against the active site of the C-terminal domain and inhibits methylesterase activity by directly restricting access to the active site. Also included in this family is chemotaxis response regulator CheY, which contains a stand-alone REC domain, and an uncharacterized subfamily composed of proteins containing an N-terminal REC domain and a C-terminal CheY-P phosphatase (CheC) domain. REC domains function as phosphorylation-mediated switches within response regulators, but some also transfer phosphoryl groups in multistep phosphorelays. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SEI00642.1 | 5.72e-240 | 29 | 486 | 35 | 474 |
| QHV64416.1 | 4.63e-212 | 20 | 487 | 27 | 481 |
| QHV71784.1 | 4.63e-212 | 20 | 487 | 27 | 481 |
| QHV69330.1 | 4.63e-212 | 20 | 487 | 27 | 481 |
| QHV66866.1 | 4.63e-212 | 20 | 487 | 27 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6T2B_A | 2.52e-192 | 25 | 486 | 6 | 447 | Glycosidehydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_B Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_C Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_D Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila] |
| 2IXA_A | 5.39e-73 | 53 | 479 | 15 | 433 | A-zyme,N-acetylgalactosaminidase [Elizabethkingia meningoseptica],2IXB_A Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC [Elizabethkingia meningoseptica] |
| 3E18_A | 3.94e-18 | 59 | 327 | 3 | 254 | CRYSTALSTRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua],3E18_B CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM Listeria innocua [Listeria innocua] |
| 3EC7_A | 6.56e-10 | 62 | 216 | 24 | 174 | CrystalStructure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_B Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_C Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_D Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_E Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_F Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_G Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_H Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 3IP3_A | 2.48e-08 | 96 | 213 | 37 | 150 | Structureof putative oxidoreductase (TM_0425) from Thermotoga maritima [Thermotoga maritima],3IP3_B Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima [Thermotoga maritima],3IP3_C Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima [Thermotoga maritima],3IP3_D Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima [Thermotoga maritima],3IP3_E Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima [Thermotoga maritima],3IP3_F Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima [Thermotoga maritima],3IP3_G Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima [Thermotoga maritima],3IP3_H Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2UL75 | 5.32e-212 | 20 | 487 | 27 | 481 | Glycosyl hydrolase family 109 protein 1 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0017 PE=3 SV=1 |
| B2UQL7 | 2.51e-191 | 25 | 486 | 31 | 472 | Glycosyl hydrolase family 109 protein 2 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0920 PE=1 SV=1 |
| Q8ECL7 | 9.77e-163 | 36 | 486 | 29 | 457 | Alpha-N-acetylgalactosaminidase OS=Shewanella oneidensis (strain MR-1) OX=211586 GN=nagA PE=3 SV=1 |
| A0KV43 | 5.59e-162 | 36 | 486 | 29 | 457 | Glycosyl hydrolase family 109 protein 1 OS=Shewanella sp. (strain ANA-3) OX=94122 GN=Shewana3_1428 PE=3 SV=1 |
| Q0HKG4 | 7.92e-162 | 36 | 486 | 29 | 457 | Glycosyl hydrolase family 109 protein 1 OS=Shewanella sp. (strain MR-4) OX=60480 GN=Shewmr4_1375 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
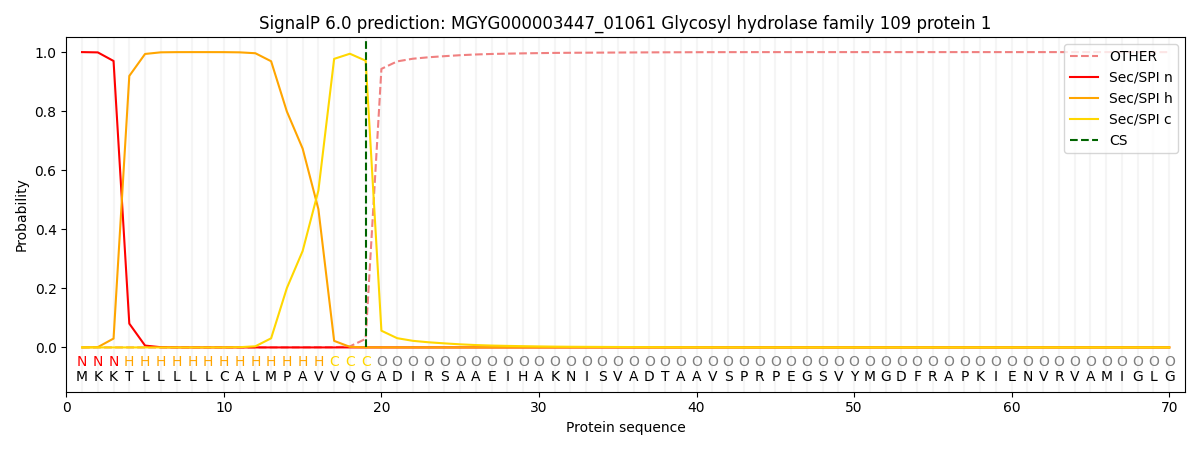
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000377 | 0.998889 | 0.000204 | 0.000182 | 0.000171 | 0.000153 |
