You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003483_00567
You are here: Home > Sequence: MGYG000003483_00567
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RUG572 sp900547945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; RUG572; RUG572 sp900547945 | |||||||||||
| CAZyme ID | MGYG000003483_00567 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10988; End: 12484 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 33 | 246 | 5.5e-50 | 0.759581881533101 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2730 | BglC | 2.91e-11 | 28 | 165 | 49 | 194 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 0.002 | 35 | 230 | 2 | 204 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDV23784.1 | 7.44e-78 | 30 | 498 | 42 | 515 |
| ACB74275.1 | 1.21e-56 | 30 | 498 | 44 | 402 |
| QPV61938.1 | 7.82e-53 | 30 | 498 | 10 | 326 |
| AQG82496.1 | 2.07e-50 | 26 | 498 | 33 | 360 |
| ARA93352.1 | 8.12e-50 | 27 | 498 | 26 | 348 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7EC9_A | 2.05e-39 | 30 | 498 | 19 | 341 | ChainA, Endoglucanase [Thermotoga maritima MSB8],7EC9_B Chain B, Endoglucanase [Thermotoga maritima MSB8],7EFZ_A Chain A, Endoglucanase [Thermotoga maritima MSB8],7EFZ_B Chain B, Endoglucanase [Thermotoga maritima MSB8] |
| 1VJZ_A | 3.74e-37 | 30 | 498 | 19 | 341 | Crystalstructure of Endoglucanase (TM1752) from Thermotoga maritima at 2.05 A resolution [Thermotoga maritima] |
| 3W0K_A | 8.22e-20 | 33 | 495 | 5 | 325 | CrystalStructure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus],3W0K_B Crystal Structure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
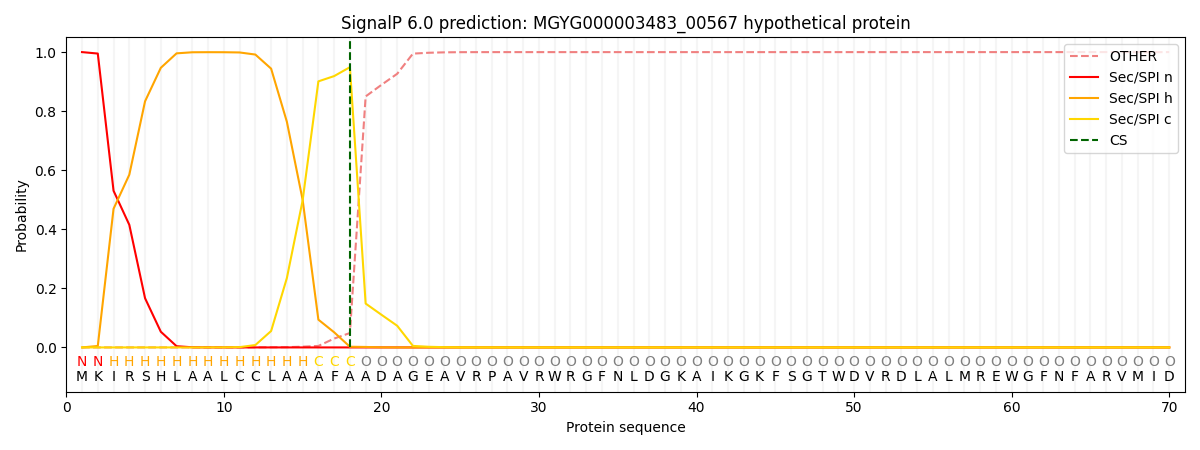
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000345 | 0.998789 | 0.000197 | 0.000234 | 0.000219 | 0.000182 |
