You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003627_01060
You are here: Home > Sequence: MGYG000003627_01060
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
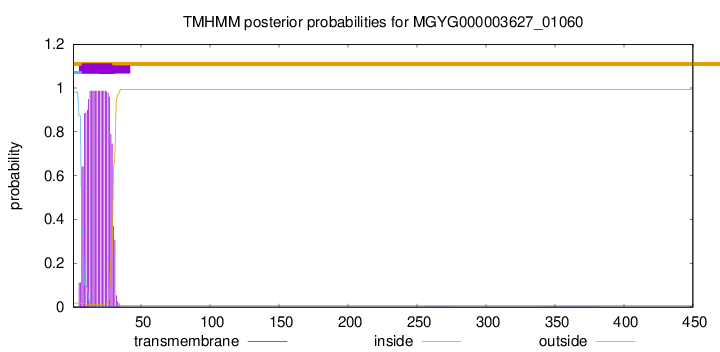
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1732; | |||||||||||
| CAZyme ID | MGYG000003627_01060 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | Multidomain esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5979; End: 7331 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 53 | 404 | 4.3e-77 | 0.9739776951672863 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAB55348.1 | 8.30e-153 | 41 | 436 | 370 | 762 |
| QTE69547.1 | 4.60e-90 | 12 | 432 | 5 | 430 |
| QTE70682.1 | 2.96e-84 | 40 | 437 | 1 | 411 |
| QTE74645.1 | 2.96e-84 | 40 | 437 | 1 | 411 |
| QUA54463.1 | 1.30e-82 | 40 | 433 | 1 | 410 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NN3_A | 4.68e-76 | 53 | 435 | 25 | 388 | ChainA, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_B Chain B, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_C Chain C, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_D Chain D, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B] |
| 6RV8_A | 1.75e-31 | 30 | 389 | 78 | 403 | CrystalStructure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
| 6RU2_A | 2.20e-31 | 31 | 389 | 1 | 325 | CrystalStructure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor],6RU2_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor] |
| 6RTV_A | 5.52e-31 | 31 | 389 | 7 | 331 | CrystalStructure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RTV_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RU1_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RU1_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RV7_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV7_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV9_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor],6RV9_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor] |
| 3PIC_A | 4.64e-30 | 28 | 405 | 4 | 343 | ChainA, Cip2 [Trichoderma reesei],3PIC_B Chain B, Cip2 [Trichoderma reesei],3PIC_C Chain C, Cip2 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9RLB8 | 1.66e-153 | 41 | 436 | 370 | 762 | Multidomain esterase OS=Ruminococcus flavefaciens OX=1265 GN=cesA PE=1 SV=1 |
| A0A0A7EQR3 | 8.87e-31 | 30 | 389 | 94 | 419 | 4-O-methyl-glucuronoyl methylesterase OS=Cerrena unicolor OX=90312 PE=1 SV=1 |
| P0CU53 | 7.89e-29 | 53 | 378 | 46 | 342 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Wolfiporia cocos (strain MD-104) OX=742152 GN=WOLCODRAFT_23632 PE=1 SV=1 |
| P0CT87 | 1.72e-28 | 53 | 378 | 112 | 407 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Phanerochaete chrysosporium (strain RP-78 / ATCC MYA-4764 / FGSC 9002) OX=273507 GN=e_gw1.18.61.1 PE=1 SV=1 |
| G0RV93 | 2.07e-28 | 28 | 405 | 89 | 428 | 4-O-methyl-glucuronoyl methylesterase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cip2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
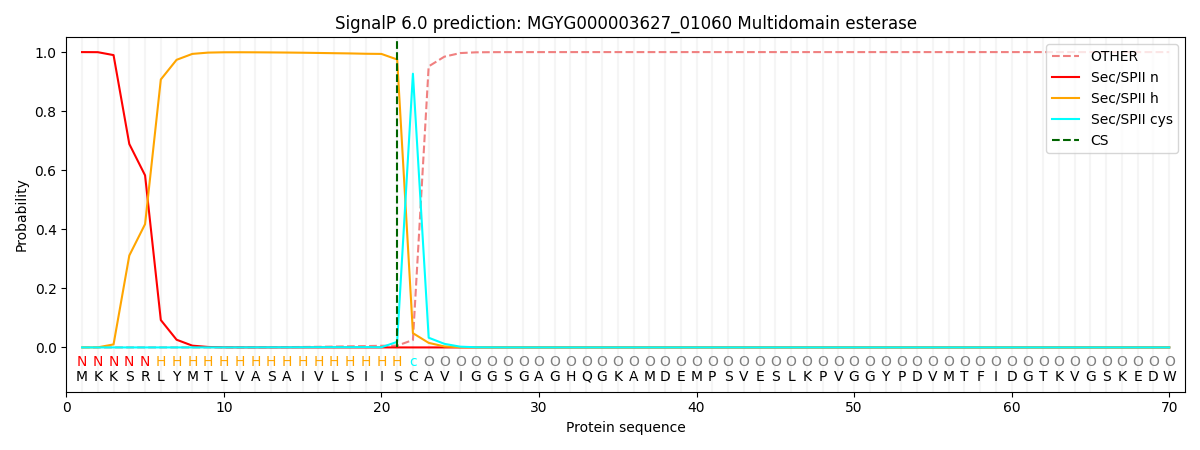
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000013 | 0.000040 | 0.999980 | 0.000000 | 0.000000 | 0.000000 |