You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003642_00070
You are here: Home > Sequence: MGYG000003642_00070
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RUG572 sp900771305 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Kiritimatiellae; RFP12; UBA1067; RUG572; RUG572 sp900771305 | |||||||||||
| CAZyme ID | MGYG000003642_00070 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6328; End: 7917 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 259 | 479 | 6.2e-33 | 0.6666666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 5.74e-26 | 114 | 479 | 3 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 7.18e-25 | 94 | 478 | 24 | 307 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.36e-16 | 137 | 479 | 69 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQZ02681.1 | 1.77e-173 | 15 | 519 | 13 | 485 |
| AHF92621.1 | 2.97e-169 | 25 | 519 | 16 | 481 |
| AWI10666.1 | 6.12e-166 | 27 | 520 | 1 | 471 |
| QGA28189.1 | 5.54e-164 | 1 | 529 | 4 | 504 |
| AVM47074.1 | 8.21e-163 | 25 | 519 | 16 | 486 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 3.32e-11 | 63 | 518 | 73 | 401 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 9.90e-10 | 63 | 518 | 73 | 401 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 3W27_A | 3.78e-09 | 269 | 482 | 125 | 326 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W28_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W29_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W24_A | 6.71e-09 | 269 | 482 | 125 | 326 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 7CPK_A | 2.35e-08 | 117 | 368 | 27 | 229 | XylanaseR from Bacillus sp. TAR-1 [Bacillus sp. TAR1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DH97 | 4.39e-08 | 255 | 515 | 500 | 733 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| P36917 | 8.72e-08 | 218 | 481 | 429 | 674 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| P07528 | 1.46e-07 | 117 | 368 | 71 | 273 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| P10478 | 2.17e-06 | 269 | 483 | 626 | 834 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| Q60041 | 3.70e-06 | 269 | 490 | 134 | 346 | Endo-1,4-beta-xylanase B OS=Thermotoga neapolitana OX=2337 GN=xynB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
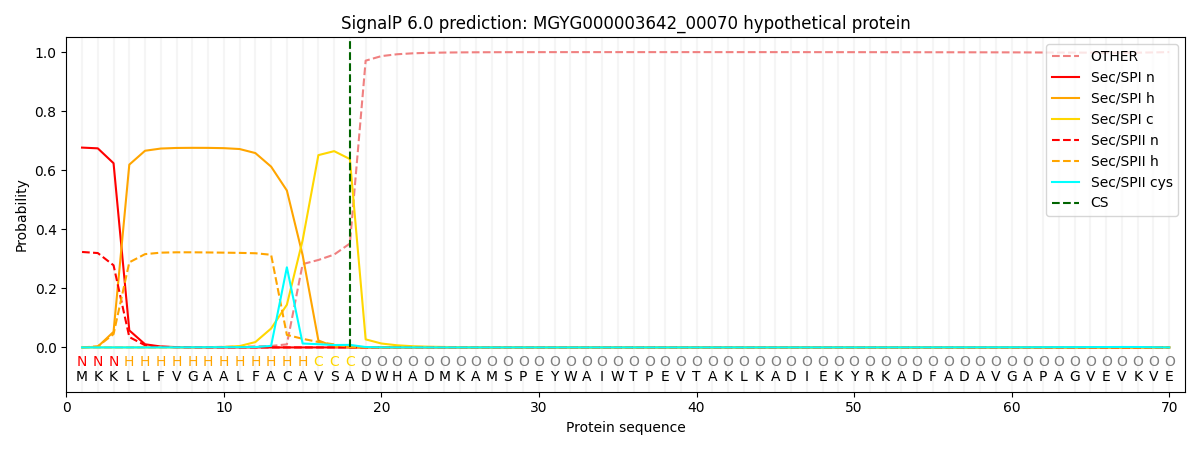
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001075 | 0.664949 | 0.333185 | 0.000266 | 0.000258 | 0.000252 |
