You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003846_01290
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312;
|
| CAZyme ID |
MGYG000003846_01290
|
| CAZy Family |
GH28 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 376 |
|
41116.05 |
8.2202 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000003846 |
2141960 |
MAG |
United States |
North America |
|
| Gene Location |
Start: 38208;
End: 39338
Strand: +
|
No EC number prediction in MGYG000003846_01290.
| Family |
Start |
End |
Evalue |
family coverage |
| GH28 |
85 |
332 |
2.4e-19 |
0.6123076923076923 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG5434
|
Pgu1 |
1.77e-11 |
32 |
297 |
98 |
409 |
Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam13229
|
Beta_helix |
1.24e-04 |
145 |
273 |
2 |
119 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| AWO01000.1
|
9.11e-119 |
21 |
356 |
5 |
333 |
| APZ82864.1
|
9.56e-28 |
21 |
365 |
26 |
349 |
This protein is predicted as SP
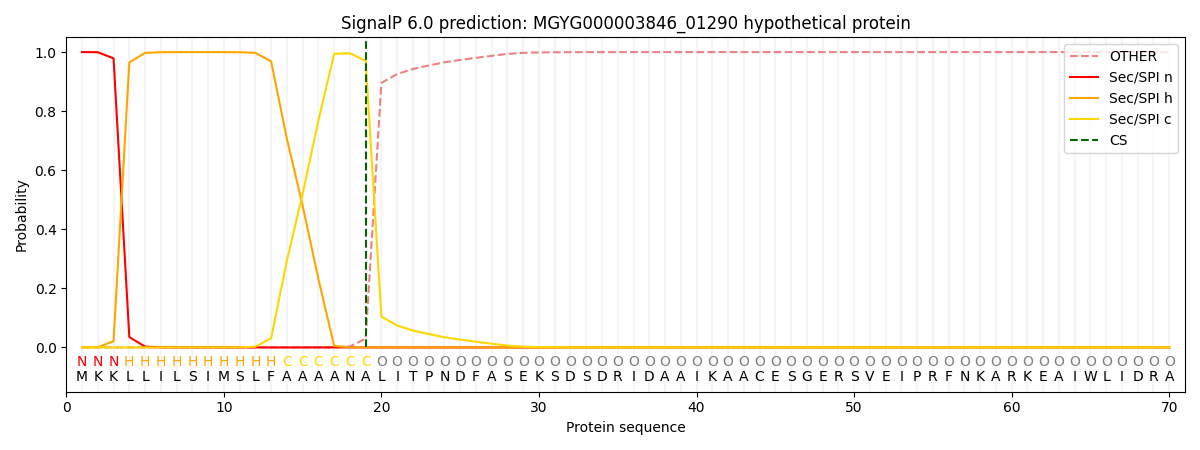
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000298
|
0.999021
|
0.000167
|
0.000185
|
0.000168
|
0.000155
|
There is no transmembrane helices in MGYG000003846_01290.

