You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004055_00465
You are here: Home > Sequence: MGYG000004055_00465
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
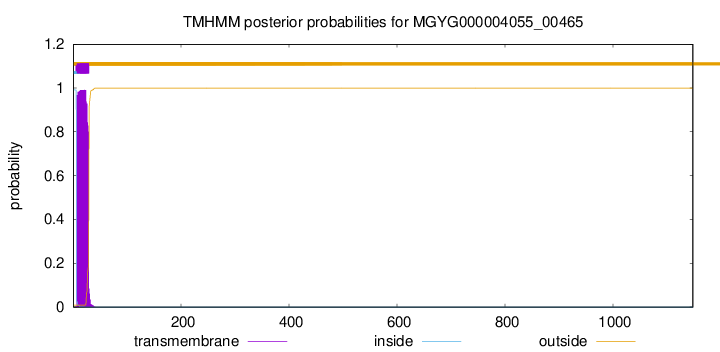
TMHMM annotations
Basic Information help
| Species | Eubacterium_G sp900548465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_G; Eubacterium_G sp900548465 | |||||||||||
| CAZyme ID | MGYG000004055_00465 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 89167; End: 92613 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 92 | 355 | 1.9e-116 | 0.9961977186311787 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.54e-26 | 115 | 354 | 26 | 268 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.54e-18 | 111 | 386 | 71 | 381 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd00257 | Fascin | 1.29e-07 | 1026 | 1145 | 10 | 119 | Fascin-like domain; members include actin-bundling/crosslinking proteins facsin, histoactophilin and singed; identified in sea urchin, Drosophila, Xenopus, rodents, and humans; The fascin-like domain adopts a beta-trefoil topology and contains an internal threefold repeat; the fascin subgroup contains four copies of the domain; Structurally similar to fibroblast growth factor (FGF) |
| cd00257 | Fascin | 1.34e-07 | 553 | 672 | 10 | 119 | Fascin-like domain; members include actin-bundling/crosslinking proteins facsin, histoactophilin and singed; identified in sea urchin, Drosophila, Xenopus, rodents, and humans; The fascin-like domain adopts a beta-trefoil topology and contains an internal threefold repeat; the fascin subgroup contains four copies of the domain; Structurally similar to fibroblast growth factor (FGF) |
| cd00257 | Fascin | 2.72e-06 | 789 | 912 | 5 | 117 | Fascin-like domain; members include actin-bundling/crosslinking proteins facsin, histoactophilin and singed; identified in sea urchin, Drosophila, Xenopus, rodents, and humans; The fascin-like domain adopts a beta-trefoil topology and contains an internal threefold repeat; the fascin subgroup contains four copies of the domain; Structurally similar to fibroblast growth factor (FGF) |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS15177.1 | 3.62e-212 | 5 | 677 | 9 | 564 |
| VEI36204.1 | 2.68e-191 | 37 | 675 | 39 | 550 |
| BCJ97694.1 | 1.40e-120 | 35 | 616 | 39 | 519 |
| AIC93076.1 | 3.02e-120 | 32 | 407 | 19 | 396 |
| QAA34430.1 | 2.89e-105 | 23 | 418 | 14 | 417 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3SO1_A | 4.67e-18 | 684 | 776 | 5 | 97 | Crystalstructure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_B Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_C Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_D Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_E Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_F Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_G Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO1_H Crystal structure of a double mutant T41S T82S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3SO0_A | 1.19e-17 | 684 | 776 | 5 | 97 | Crystalstructure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_B Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_C Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_D Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_E Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_F Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_G Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3SO0_H Crystal structure of a mutant T41S of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3SNZ_A | 2.21e-17 | 684 | 776 | 5 | 97 | Crystalstructure of a mutant W39D of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 3IAJ_A | 3.10e-17 | 684 | 766 | 2 | 84 | Crystalstructure of a betagamma-crystallin domain from Clostridium beijerinckii-in alternate space group I422 [Clostridium beijerinckii NCIMB 8052] |
| 3I9H_A | 3.19e-17 | 684 | 766 | 3 | 85 | Crystalstructure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_B Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_C Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_D Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_E Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_F Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_G Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052],3I9H_H Crystal structure of a betagamma-crystallin domain from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W8QRE4 | 2.41e-25 | 43 | 308 | 3 | 264 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| B8N151 | 2.39e-18 | 57 | 262 | 27 | 232 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=exgA PE=3 SV=1 |
| Q7Z9L3 | 2.39e-18 | 57 | 262 | 27 | 232 | Glucan 1,3-beta-glucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=exgA PE=1 SV=1 |
| B0XN12 | 2.67e-18 | 63 | 263 | 42 | 245 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
| Q4WK60 | 3.57e-18 | 63 | 256 | 42 | 238 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
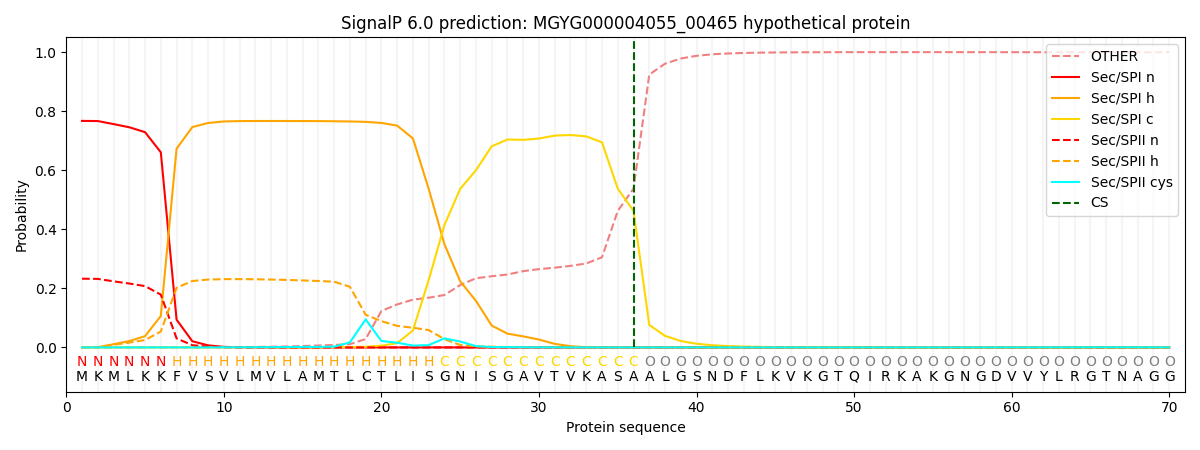
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000886 | 0.761358 | 0.237013 | 0.000266 | 0.000229 | 0.000221 |