You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004210_00048
You are here: Home > Sequence: MGYG000004210_00048
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
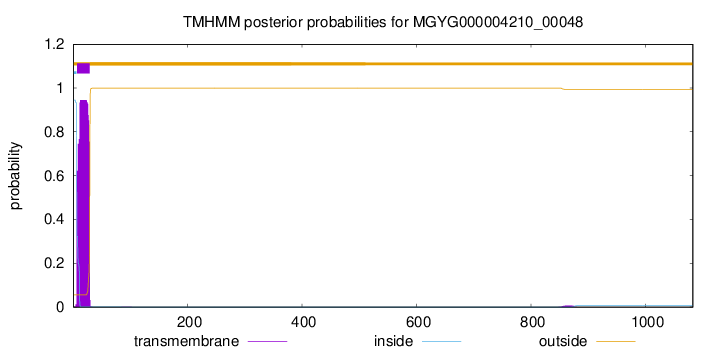
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; V9D3004; | |||||||||||
| CAZyme ID | MGYG000004210_00048 | |||||||||||
| CAZy Family | GH59 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45839; End: 49090 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH59 | 56 | 738 | 3.9e-153 | 0.9920760697305864 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02057 | Glyco_hydro_59 | 4.13e-85 | 61 | 397 | 1 | 291 | Glycosyl hydrolase family 59. |
| COG5492 | YjdB | 8.08e-12 | 837 | 985 | 145 | 287 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam06439 | DUF1080 | 5.31e-08 | 581 | 738 | 17 | 182 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
| COG5520 | XynC | 1.27e-07 | 146 | 306 | 110 | 270 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02368 | Big_2 | 1.73e-07 | 879 | 944 | 8 | 72 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMW11854.1 | 2.34e-254 | 21 | 919 | 19 | 958 |
| QLH25411.1 | 2.27e-251 | 2 | 922 | 5 | 961 |
| QUW95523.1 | 2.26e-248 | 21 | 860 | 22 | 906 |
| AZM74018.1 | 9.14e-247 | 37 | 860 | 34 | 903 |
| QKW59509.1 | 7.24e-246 | 37 | 860 | 34 | 903 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5SNX7 | 4.40e-17 | 65 | 709 | 38 | 628 | Galactocerebrosidase OS=Danio rerio OX=7955 GN=galc PE=2 SV=1 |
| Q0VA39 | 1.25e-15 | 40 | 649 | 24 | 576 | Galactocerebrosidase OS=Xenopus tropicalis OX=8364 GN=galc PE=2 SV=1 |
| Q498K0 | 1.42e-11 | 58 | 649 | 44 | 575 | Galactocerebrosidase OS=Xenopus laevis OX=8355 GN=galc PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
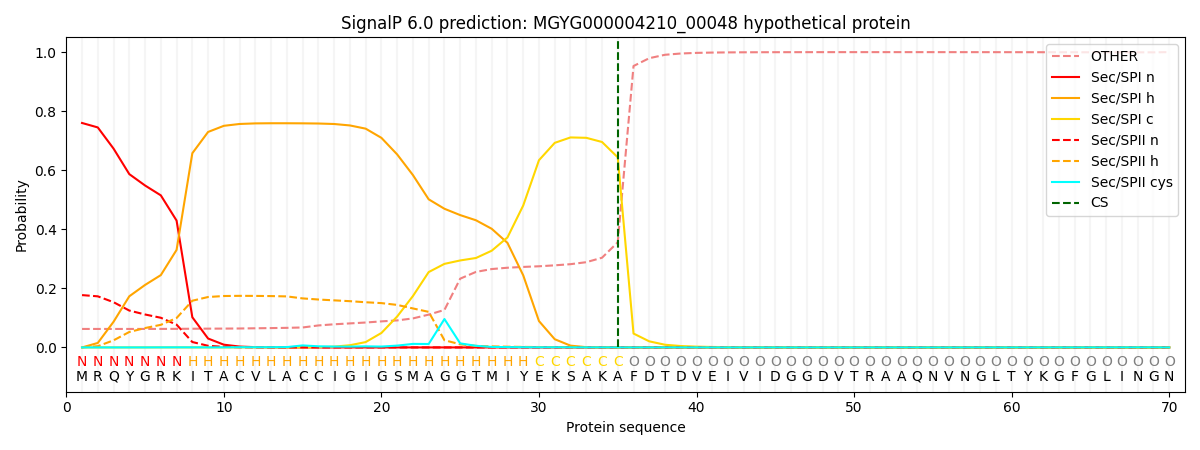
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.065612 | 0.754130 | 0.179008 | 0.000697 | 0.000272 | 0.000249 |