You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004380_02146
You are here: Home > Sequence: MGYG000004380_02146
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000004380_02146 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13954; End: 16446 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.41e-21 | 483 | 716 | 41 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 1.31e-08 | 318 | 373 | 1 | 54 | Bacterial SH3 domain. |
| pfam08239 | SH3_3 | 5.69e-08 | 52 | 111 | 1 | 54 | Bacterial SH3 domain. |
| pfam08239 | SH3_3 | 5.69e-07 | 126 | 181 | 1 | 54 | Bacterial SH3 domain. |
| pfam08239 | SH3_3 | 4.05e-06 | 230 | 285 | 1 | 54 | Bacterial SH3 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK83300.1 | 1.03e-110 | 159 | 829 | 11 | 631 |
| QNM00733.1 | 3.07e-108 | 342 | 829 | 70 | 528 |
| BCJ93456.1 | 1.89e-106 | 17 | 830 | 11 | 895 |
| QWT53775.1 | 7.20e-105 | 342 | 829 | 70 | 528 |
| AEN96074.1 | 1.37e-104 | 213 | 829 | 34 | 611 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PI7_A | 8.40e-09 | 509 | 708 | 37 | 222 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 2KYB_A | 4.56e-08 | 222 | 284 | 4 | 60 | Solutionstructure of CpR82G from Clostridium perfringens. North East Structural Genomics Consortium Target CpR82g [Clostridium perfringens ATCC 13124] |
| 4PI8_A | 5.01e-08 | 509 | 708 | 37 | 222 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 2KT8_A | 5.08e-08 | 219 | 284 | 2 | 61 | SolutionNMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B [Clostridium perfringens] |
| 6FXO_A | 1.15e-06 | 608 | 715 | 149 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CPQ1 | 8.07e-08 | 490 | 715 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 1.83e-07 | 490 | 715 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 1.83e-07 | 490 | 715 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
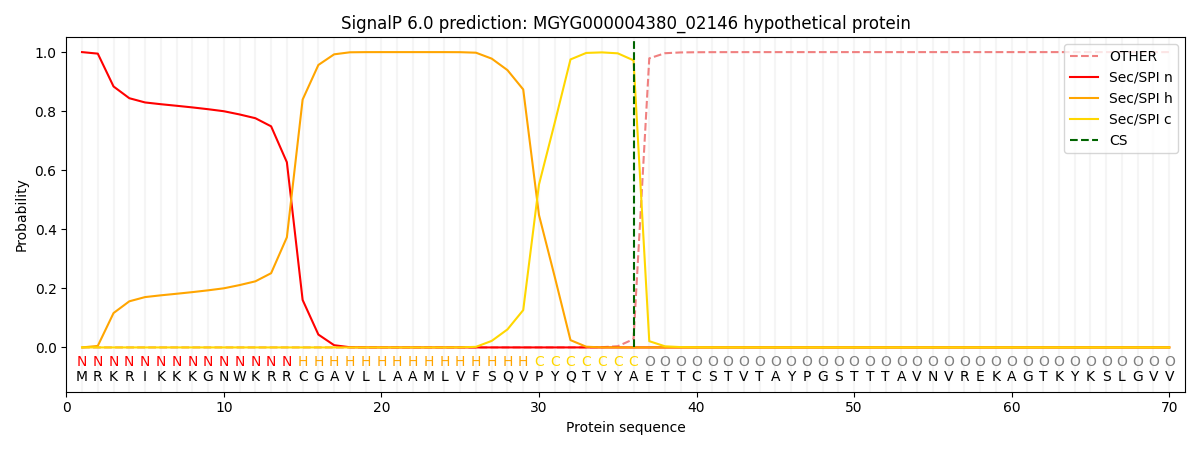
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000403 | 0.998869 | 0.000194 | 0.000195 | 0.000165 | 0.000150 |
