You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004462_00741
You are here: Home > Sequence: MGYG000004462_00741
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004462_00741 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 68090; End: 72592 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 1050 | 1308 | 2.7e-44 | 0.8541666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 5.04e-19 | 1019 | 1288 | 58 | 358 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 1.04e-17 | 1050 | 1294 | 14 | 248 | Pectinesterase. |
| PLN02708 | PLN02708 | 4.90e-17 | 1050 | 1374 | 255 | 553 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02217 | PLN02217 | 1.25e-15 | 1042 | 1455 | 251 | 667 | probable pectinesterase/pectinesterase inhibitor |
| PLN02497 | PLN02497 | 2.56e-15 | 1169 | 1290 | 149 | 272 | probable pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD41145.1 | 0.0 | 1 | 1498 | 1 | 1492 |
| QCD38476.1 | 0.0 | 11 | 1498 | 10 | 1473 |
| QCP72166.1 | 0.0 | 11 | 1498 | 10 | 1473 |
| QNT65238.1 | 0.0 | 403 | 1498 | 24 | 1130 |
| QUT74167.1 | 0.0 | 11 | 1497 | 9 | 1431 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 3.28e-15 | 1036 | 1293 | 26 | 308 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 1XG2_A | 1.17e-13 | 1047 | 1345 | 14 | 293 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 4PMH_A | 1.44e-12 | 1028 | 1316 | 21 | 352 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
| 2NSP_A | 4.98e-12 | 1169 | 1372 | 139 | 340 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1QJV_A | 2.08e-09 | 1169 | 1372 | 139 | 340 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q1JPL7 | 2.54e-15 | 1031 | 1335 | 235 | 534 | Pectinesterase/pectinesterase inhibitor 18 OS=Arabidopsis thaliana OX=3702 GN=PME18 PE=1 SV=3 |
| Q9FJ21 | 2.23e-13 | 1050 | 1317 | 272 | 530 | Probable pectinesterase/pectinesterase inhibitor 58 OS=Arabidopsis thaliana OX=3702 GN=PME58 PE=2 SV=1 |
| P14280 | 2.52e-12 | 1047 | 1345 | 243 | 522 | Pectinesterase 1 OS=Solanum lycopersicum OX=4081 GN=PME1.9 PE=1 SV=5 |
| P09607 | 1.01e-11 | 1050 | 1328 | 250 | 518 | Pectinesterase 2.1 OS=Solanum lycopersicum OX=4081 GN=PME2.1 PE=2 SV=2 |
| Q43043 | 1.49e-11 | 1050 | 1288 | 71 | 299 | Pectinesterase OS=Petunia integrifolia OX=4103 GN=PPE1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
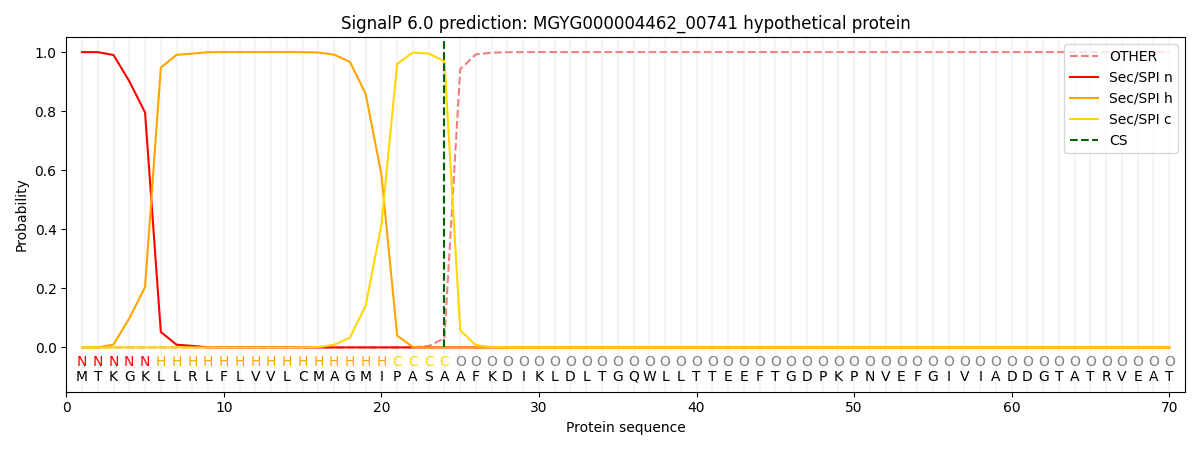
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000342 | 0.998795 | 0.000345 | 0.000178 | 0.000169 | 0.000153 |

