You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004499_01668
You are here: Home > Sequence: MGYG000004499_01668
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp900551245 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900551245 | |||||||||||
| CAZyme ID | MGYG000004499_01668 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14069; End: 15934 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 109 | 260 | 1.4e-22 | 0.39675174013921116 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1874 | GanA | 9.65e-07 | 26 | 210 | 18 | 229 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam02449 | Glyco_hydro_42 | 6.37e-06 | 41 | 126 | 13 | 125 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| pfam11790 | Glyco_hydro_cc | 1.98e-04 | 102 | 246 | 44 | 174 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| COG2723 | BglB | 0.002 | 41 | 142 | 62 | 174 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| PRK15014 | PRK15014 | 0.005 | 41 | 146 | 72 | 188 | 6-phospho-beta-glucosidase BglA; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 4.43e-153 | 4 | 620 | 3 | 619 |
| QDU57372.1 | 4.55e-63 | 8 | 618 | 7 | 631 |
| AHF90941.1 | 8.54e-39 | 4 | 580 | 203 | 814 |
| AOD20726.1 | 4.82e-33 | 38 | 251 | 54 | 287 |
| QXU53077.1 | 1.23e-32 | 38 | 251 | 54 | 287 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZN2_A | 2.18e-09 | 48 | 244 | 43 | 254 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 2.18e-09 | 48 | 244 | 43 | 254 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
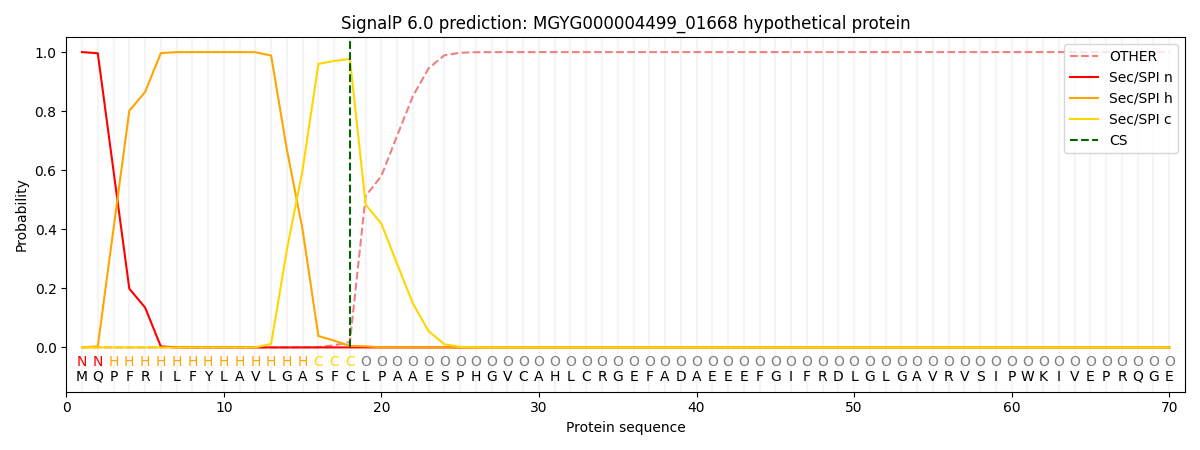
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000195 | 0.999193 | 0.000149 | 0.000151 | 0.000138 | 0.000132 |
